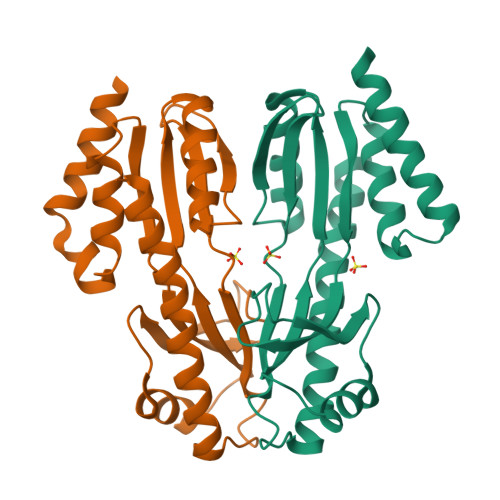
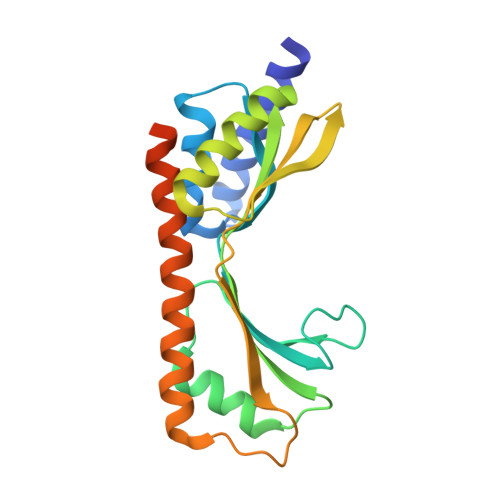
Structural and functional characterization of the TYW3/Taw3 class of SAM-dependent methyltransferases.
Currie, M.A., Brown, G., Wong, A., Ohira, T., Sugiyama, K., Suzuki, T., Yakunin, A.F., Jia, Z.(2017) RNA 23: 346-354
- PubMed: 27932585
- DOI: https://doi.org/10.1261/rna.057943.116
- Primary Citation of Related Structures:
1TLJ - PubMed Abstract:
S -adenosylmethionine (SAM)-dependent methyltransferases regulate a wide range of biological processes through the modification of proteins, nucleic acids, polysaccharides, as well as various metabolites. TYW3/Taw3 is a SAM-dependent methyltransferase responsible for the formation of a tRNA modification known as wybutosine and its derivatives that are required for accurate decoding in protein synthesis. Here, we report the crystal structure of Taw3, a homolog of TYW3 from Sulfolobus solfataricus , which revealed a novel α/β fold. The sequence motif (S/T)xSSCxGR and invariant aspartate and histidine, conserved in TYW3/Taw3, cluster to form the catalytic center. These structural and sequence features indicate that TYW3/Taw3 proteins constitute a distinct class of SAM-dependent methyltransferases. Using site-directed mutagenesis along with in vivo complementation assays combined with mass spectrometry as well as ligand docking and cofactor binding assays, we have identified the active site of TYW3 and residues essential for cofactor binding and methyltransferase activity.
Organizational Affiliation:
Department of Biomedical and Molecular Sciences, Queen's University, Kingston, Ontario K7L 3N6, Canada.