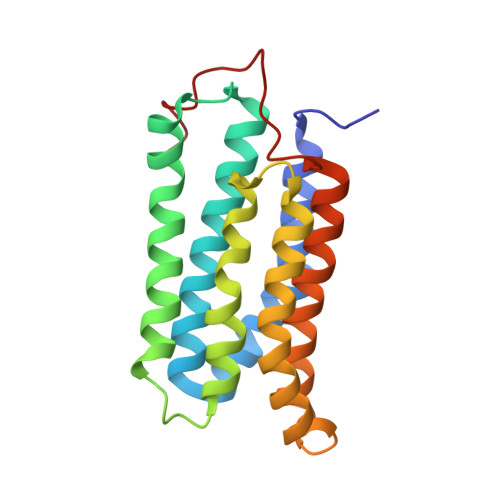
Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP).
Mohd Yusof, A., Hu, N.J., Wlodawer, A., Hofmann, A.(2005) Proteins 58: 255-262
- PubMed: 15558566
- DOI: https://doi.org/10.1002/prot.20314
- Primary Citation of Related Structures:
1TJF - PubMed Abstract:
Cyclase-associated protein (CAP) is a highly conserved and widely distributed protein that links the nutritional response signaling to cytoskeleton remodeling. In yeast, CAP is a component of the adenylyl cyclase complex and helps to activate the Ras-mediated catalytic cycle of the cyclase. While the N-terminal domain of CAP (N-CAP) provides a binding site for adenylyl cyclase, the C-terminal domain (C-CAP) possesses actin binding activity. Our attempts to crystallize full-length recombinant CAP from Dictyostelium discoideum resulted in growth of orthorhombic crystals containing only the N-terminal domain (residues 42-227) due to auto-proteolytic cleavage. The structure was solved by molecular replacement with data at 2.2 A resolution. The present crystal structure allows the characterization of a head-to-tail N-CAP dimer in the asymmetric unit and a crystallographic side-to-side dimer. Comparison with previously published structures of N-CAP reveals variable modes of dimerization of this domain, but the presence of a common interface for the side-to-side dimer.
Organizational Affiliation:
Institute of Structural & Molecular Biology, School of Biological Sciences, The University of Edinburgh, Scotland, United Kingdom.