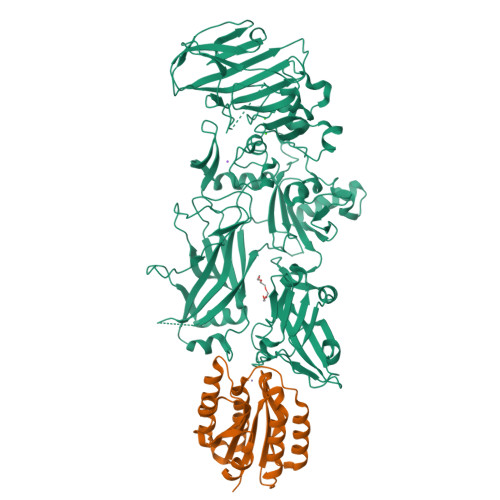
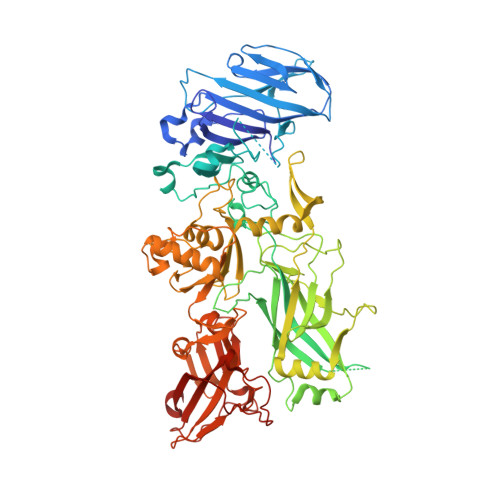
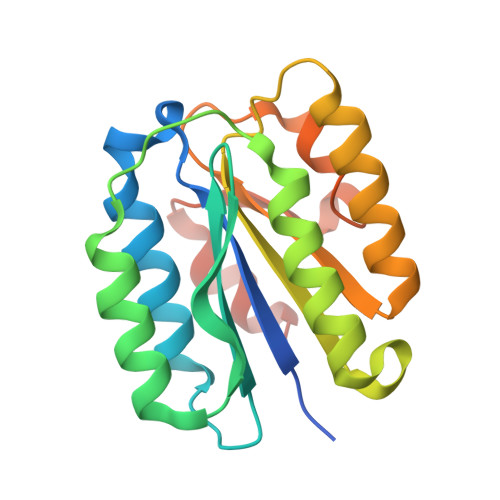
Crystal structure of a complex between anthrax toxin and its host cell receptor
Santelli, E., Bankston, L.A., Leppla, S.H., Liddington, R.C.(2004) Nature 430: 905-908
- PubMed: 15243628
- DOI: https://doi.org/10.1038/nature02763
- Primary Citation of Related Structures:
1T6B - PubMed Abstract:
Anthrax toxin consists of the proteins protective antigen (PA), lethal factor (LF) and oedema factor (EF). The first step of toxin entry into host cells is the recognition by PA of a receptor on the surface of the target cell. Subsequent cleavage of receptor-bound PA enables EF and LF to bind and form a heptameric PA63 pre-pore, which triggers endocytosis. Upon acidification of the endosome, PA63 forms a pore that inserts into the membrane and translocates EF and LF into the cytosol. Two closely related host cell receptors, TEM8 and CMG2, have been identified. Both bind to PA with high affinity and are capable of mediating toxicity. Here, we report the crystal structure of the PA-CMG2 complex at 2.5 A resolution. The structure reveals an extensive receptor-pathogen interaction surface mimicking the non-pathogenic recognition of the extracellular matrix by integrins. The binding surface is closely conserved in the two receptors and across species, but is quite different in the integrin domains, explaining the specificity of the interaction. CMG2 engages two domains of PA, and modelling of the receptor-bound PA63 heptamer suggests that the receptor acts as a pH-sensitive brace to ensure accurate and timely membrane insertion. The structure provides new leads for the discovery of anthrax anti-toxins, and should aid the design of cancer therapeutics.
Organizational Affiliation:
Program on Cell Adhesion, The Burnham Institute, 10901 North Torrey Pines Road, La Jolla, California 92037, USA.