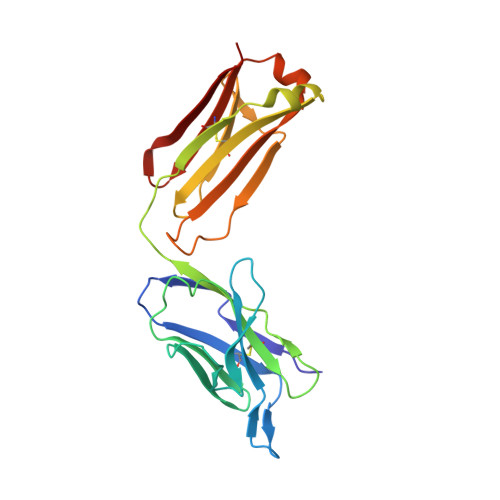
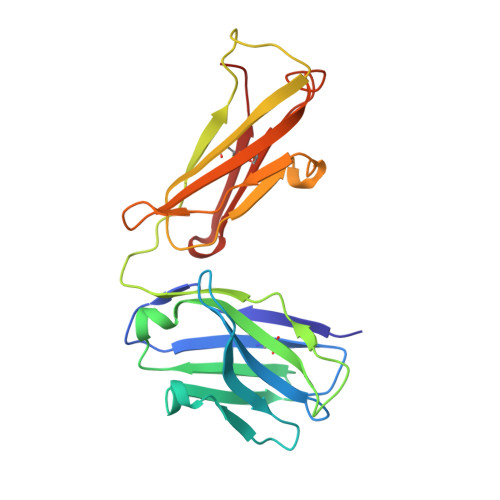
The Origin of Enantioselectivity in Aldolase Antibodies: Crystal Structure, Site-directed Mutagenesis, and Computational Analysis
Zhu, X., Tanaka, F., Hu, Y., Heine, A., Fuller, R., Zhong, G., Olson, A.J., Lerner, R.A., Barbas, C.F., Wilson, I.A.(2004) J Mol Biology 343: 1269-1280
- PubMed: 15491612
- DOI: https://doi.org/10.1016/j.jmb.2004.08.102
- Primary Citation of Related Structures:
1T4K - PubMed Abstract:
Catalytic aldolase antibodies, generated by reactive immunization, catalyze the aldol reaction with the efficiency of natural enzymes, but accept a much broader range of substrates. Two separate groups of aldolase antibodies that catalyze the same aldol reactions with antipodal selectivity were analyzed by comparing their amino acid sequences with their crystal structures, site-directed mutagenesis data, and computational docking of the transition states of the aldol reaction. The crystal structure of aldolase antibody 93F3 Fab' at 2.5A resolution revealed a combining site with two lysine residues, including LysL89 that reacts to form the covalent enamine intermediate. In contrast, antibody 33F12 has one active site lysine, LysH93. The reactive lysine residues in each group of antibodies are differentially located on the heavy and light chain variable regions in pseudo-symmetric opposite orientations, but both within highly hydrophobic environments. Thus, the defining feature for the observed enantioselectivities of these aldolase antibody catalysts is the respective location and relative disposition of the reactive lysine residues within the active sites of these catalysts.
Organizational Affiliation:
Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.