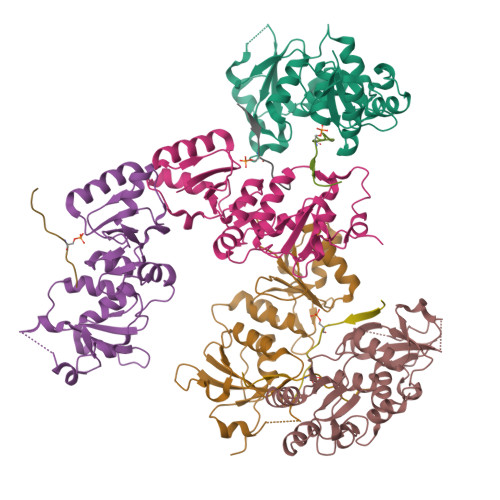
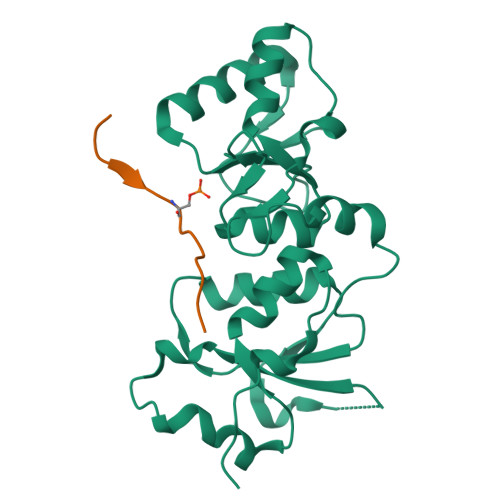
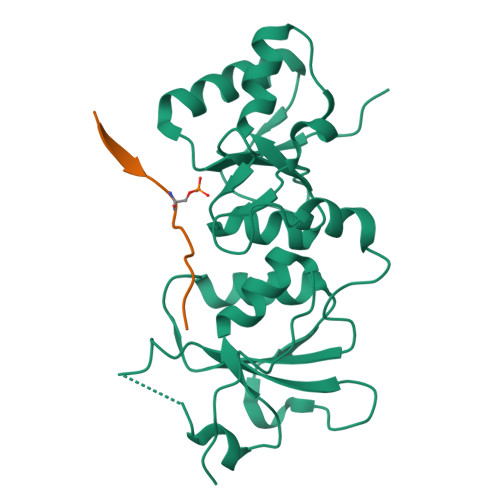
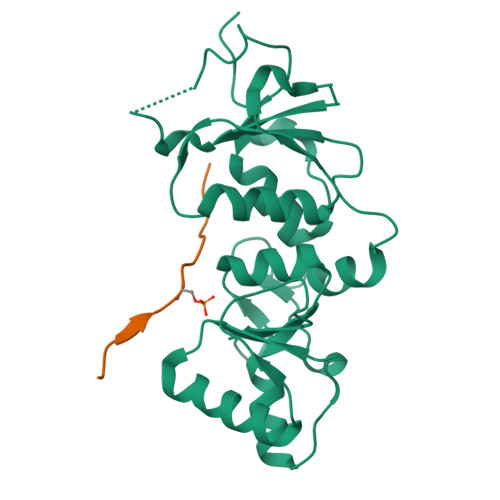
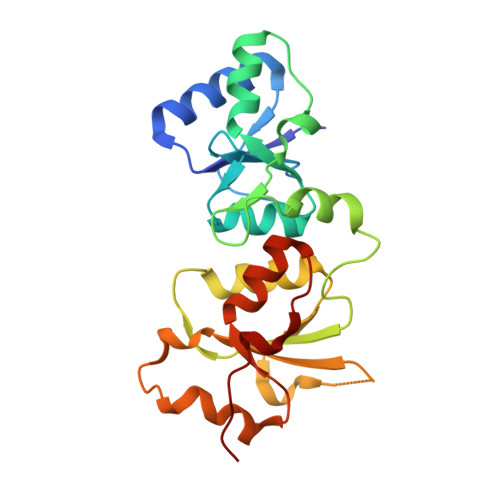
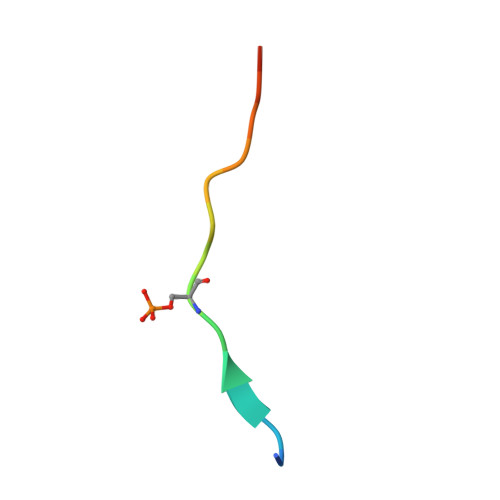
Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1
Williams, R.S., Lee, M.S., Hau, D.D., Glover, J.N.M.(2004) Nat Struct Mol Biol 11: 519-525
- PubMed: 15133503
- DOI: https://doi.org/10.1038/nsmb776
- Primary Citation of Related Structures:
1T2U, 1T2V - PubMed Abstract:
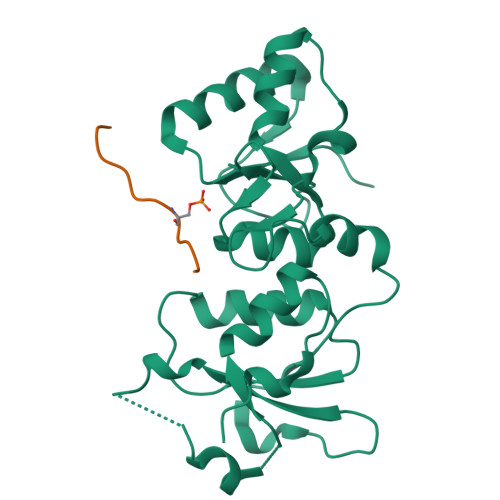
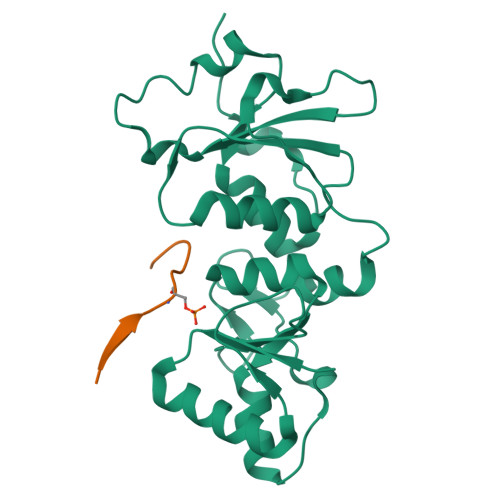
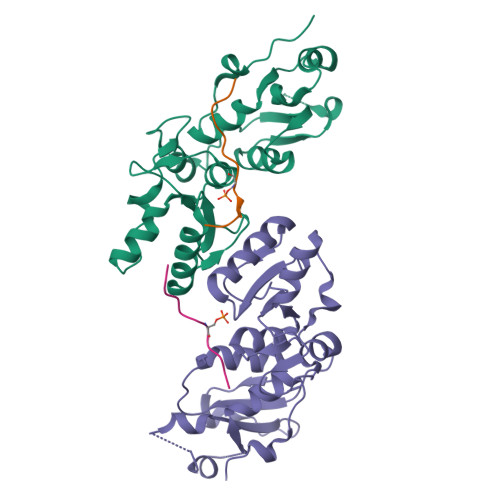
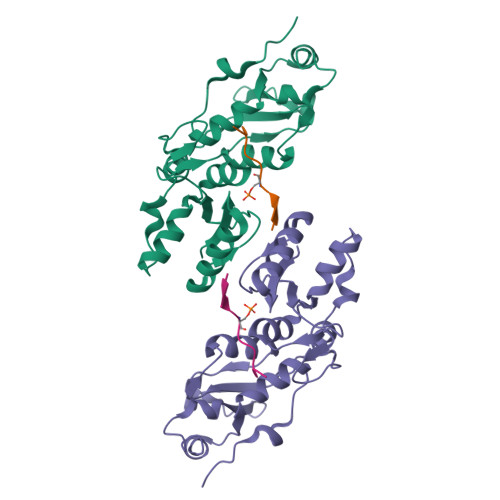
The BRCT repeats in BRCA1 are essential for its tumor suppressor activity and interact with phosphorylated protein targets containing the sequence pSer-X-X-Phe, where X indicates any residue. The structure of the tandem BRCA1 BRCT repeats bound to an optimized phosphopeptide reveals that the N-terminal repeat harbors a conserved BRCT phosphoserine-binding pocket, while the interface between the repeats forms a hydrophobic groove that recognizes the phenylalanine. Crystallographic and biochemical data suggest that the structural integrity of both binding sites is essential for peptide recognition. The diminished peptide-binding capacity observed for cancer-associated BRCA1-BRCT variants may explain the enhanced cancer risks associated with these mutations.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Alberta, T6G 2H7, Canada.