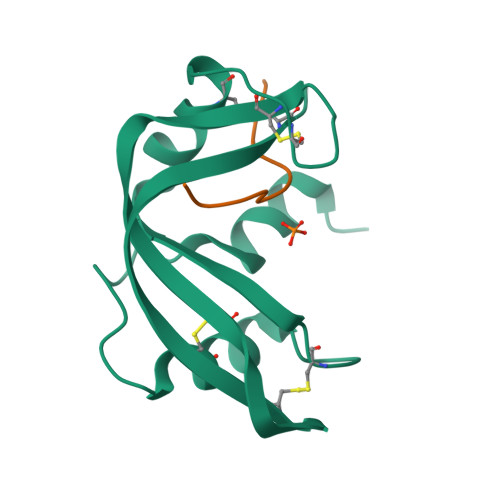
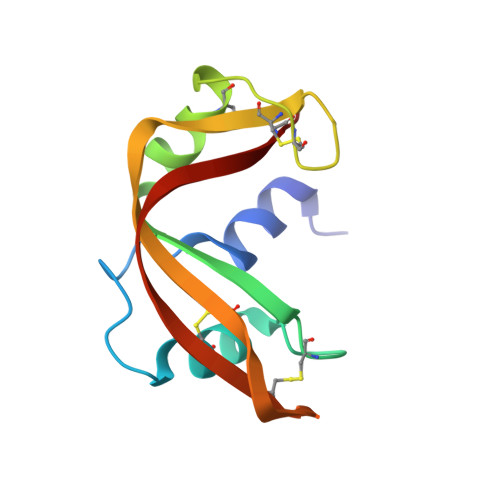
1.6 A structure of semisynthetic ribonuclease crystallized from aqueous ethanol. Comparison with crystals from salt solutions and with ribonuclease A from aqueous alcohol solutions.
de Mel, S.J., Doscher, M.S., Martin, P.D., Rodier, F., Edwards, B.F.(1995) Acta Crystallogr D Biol Crystallogr 51: 1003-1012
- PubMed: 15299768
- DOI: https://doi.org/10.1107/S0907444995004574
- Primary Citation of Related Structures:
1SSC - PubMed Abstract:
The non-covalent combination of residues 1-118 of RNase A with a synthetic 14-residue peptide containing residues 111-124 of the molecule forms a highly active semisynthetic enzyme, RNase 1-118:111-124. With this enzyme, the roles played by the six C-terminal residues in generating the catalytic efficiency and substrate specificity of RNase can be studied using chemically synthesized analogs. The structure of RNase 1-118:111-124 from 43% aqueous ethanol has been determined using molecular-replacement methods and refined to a crystallographic R-factor of 0.166 for all observed reflections in the range 7.0-1.6 A (Protein Data Bank file ISSC). The structure is compared with the 2.0 A structure of RNase A from 43% aqueous 2-methyl-2-propanol and with the 1.8 A structure of the semisynthetic enzyme obtained from crystals grown in concentrated salt solution. The structure of RNase 1-118:111-124 from aqueous ethanol is virtually identical to that of RNase A from aqueous 2-methyl-2-propanol. Half of the crystallographically bound water molecules are not coincident, however. The structure is somewhat less similar to that of RNase 1-118:111-124 from salt solutions, with a major difference being the positioning of active-site residue His119.
Organizational Affiliation:
Department of Biochemistry, Wayne State University School of Medicine, Detroit, MI 48201, USA.