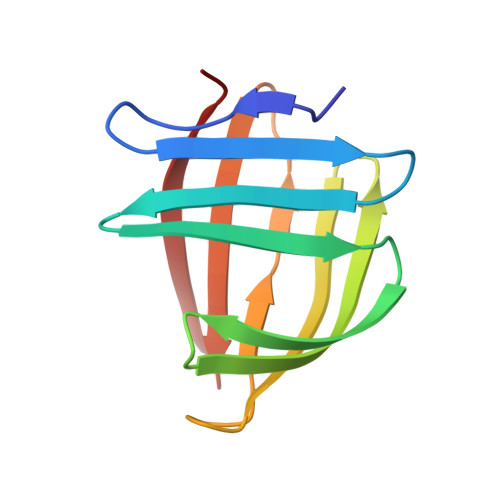
The NMR structure of a stable and compact all-beta-sheet variant of intestinal fatty acid-binding protein.
Ogbay, B., Dekoster, G.T., Cistola, D.P.(2004) Protein Sci 13: 1227-1237
- PubMed: 15096629
- DOI: https://doi.org/10.1110/ps.03546204
- Primary Citation of Related Structures:
1SA8 - PubMed Abstract:
Intestinal fatty acid-binding protein (I-FABP) has a clam-shaped structure that may serve as a scaffold for the design of artificial enzymes and drug carriers. In an attempt to optimize the scaffold for increased access to the interior-binding cavity, several helix-less variants of I-FABP have been engineered. The solution-state NMR structure of the first generation helix-less variant, known as Delta17-SG, revealed a larger-than-expected and structurally ill-defined loop flanking the deletion site. We hypothesized that the presence of this loop, on balance, was energetically unfavorable for the stability of the protein. The structure exhibited no favorable pairwise or nonpolar interactions in the loop that could offset the loss of configurational entropy associated with the folding of this region of the protein. As an attempt to generate a more stable protein, we engineered a second-generation helix-less variant of I-FABP (Delta27-GG) by deleting 27 contiguous residues of the wild-type protein and replacing them with a G-G linker. The deletion site of this variant (D9 through N35) includes the 10 residues spanning the unstructured loop of Delta17-SG. Chemical denaturation experiments using steady-state fluorescence spectroscopy showed that the second-generation helix-less variant is energetically more stable than Delta17-SG. The three-dimensional structure of apo-Delta27-GG was solved using triple-resonance NMR spectroscopy along with the structure calculation and refinement protocols contained in the program package ARIA/CNS. In spite of the deletion of 27 residues, the structure assumes a compact all-beta-sheet fold with no unstructured loops and open access to the interior cavity.
Organizational Affiliation:
Department of Chemistry, Washington University School of Medicine, St. Louis, Missouri 63110, USA.