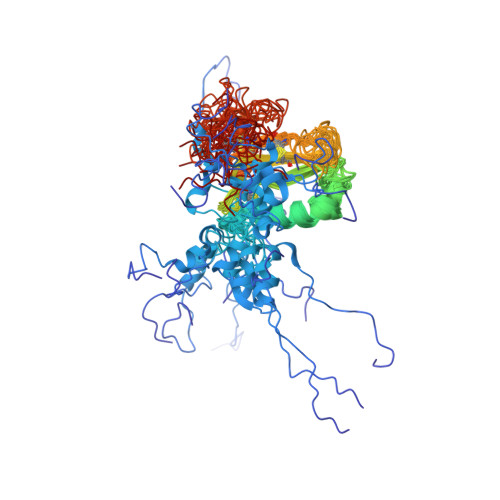
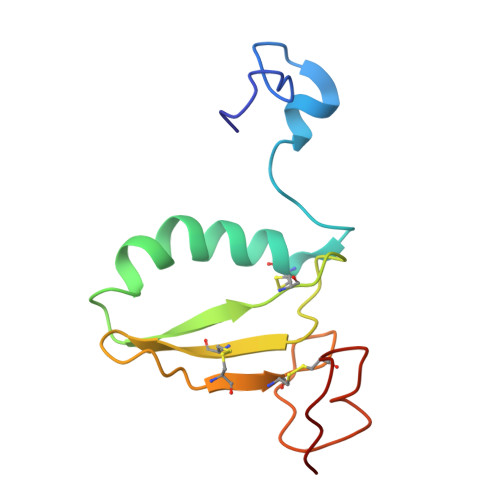
C-terminal domain of insulin-like growth factor (IGF) binding protein-6: structure and interaction with IGF-II.
Headey, S.J., Keizer, D.W., Yao, S., Brasier, G., Kantharidis, P., Bach, L.A., Norton, R.S.(2004) Mol Endocrinol 18: 2740-2750
- PubMed: 15308688
- DOI: https://doi.org/10.1210/me.2004-0248
- Primary Citation of Related Structures:
1RMJ - PubMed Abstract:
IGFs are important mediators of growth. IGF binding proteins (IGFBPs) 1-6 regulate IGF actions and have IGF-independent actions. The C-terminal domains of IGFBPs contribute to high-affinity IGF binding and modulation of IGF actions and confer some IGF-independent properties, but understanding how they achieve this has been constrained by the lack of a three-dimensional structure. We therefore determined the solution structure of the C-domain of IGFBP-6 using nuclear magnetic resonance (NMR). The domain consists of a thyroglobulin type 1 fold comprising an alpha-helix followed by a loop, a three-stranded antiparallel beta-sheet incorporating a second loop, and finally a disulfide-bonded flexible third loop. The IGF-II binding site on the C-domain was identified by examining NMR spectral changes upon complex formation. It consists of a largely hydrophobic surface patch involving the alpha-helix, the first beta-strand, and the first and second loops. The site was confirmed by mutagenesis of several residues, which resulted in decreased IGF binding affinity. The IGF-II binding site lies adjacent to surfaces likely to be involved in glycosaminoglycan binding of IGFBPs, which might explain their decreased IGF affinity when bound to glycosaminoglycans, and nuclear localization. Our structure provides a framework for understanding the roles of IGFBP C-domains in modulating IGF actions and conferring IGF-independent actions, as well as ultimately for the development of therapeutic IGF inhibitors for diseases including cancer.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville 3050, Australia.