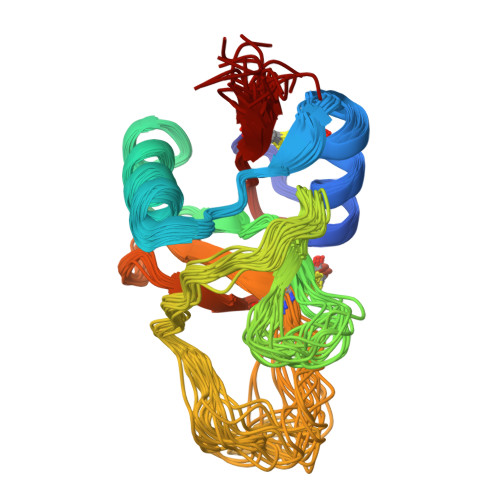
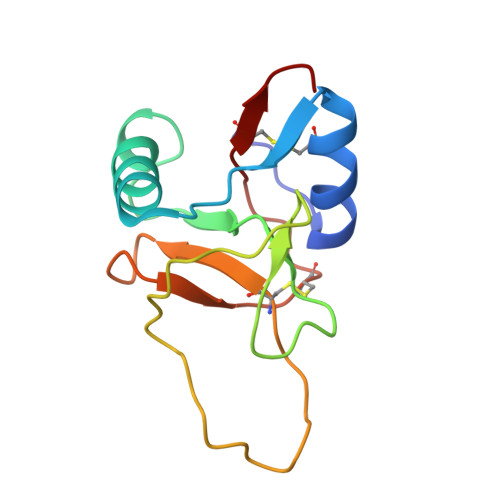
Structure of the Plasminogen Kringle 4 Binding Calcium-Free Form of the C-Type Lectin-Like Domain of Tetranectin.
Nielbo, S., Thomsen, J.K., Graversen, J.H., Jensen, P.H., Etzerodt, M., Poulsen, F.M., Thoegersen, H.C.(2004) Biochemistry 43: 8636-8643
- PubMed: 15236571
- DOI: https://doi.org/10.1021/bi049570s
- Primary Citation of Related Structures:
1RJH - PubMed Abstract:
Tetranectin is a homotrimeric protein containing a C-type lectin-like domain. This domain (TN3) can bind calcium, but in the absence of calcium, the domain binds a number of kringle-type protein ligands. Two of the calcium-coordinating residues are also critical for binding plasminogen kringle 4 (K4). The structure of the calcium free-form of TN3 (apoTN3) has been determined by NMR. Compared to the structure of the calcium-bound form of TN3 (holoTN3), the core region of secondary structural elements is conserved, while large displacements occur in the loops involved in calcium or K4 binding. A conserved proline, which was found to be in the cis conformation in holoTN3, is in apoTN3 predominantly in the trans conformation. Backbone dynamics indicate that, in apoTN3 especially, two of the three calcium-binding loops and two of the three K4-binding residues exhibit increased flexibility, whereas no such flexibility is observed in holoTN3. In the 20 best nuclear magnetic resonance structures of apoTN3, the residues critical for K4 binding span a large conformational space. Together with the relaxation data, this indicates that the K4-ligand-binding site in apoTN3 is not preformed.
Organizational Affiliation:
Laboratory of Gene Expression, Department of Molecular and Structural Biology, University of Aarhus, Gustav Wieds Vej 10, DK-8000 Aarhus C, Denmark.