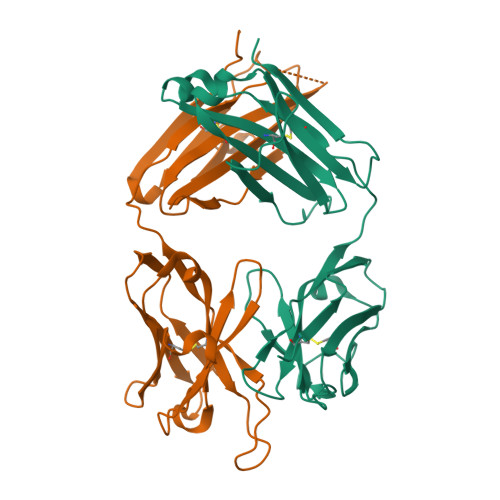
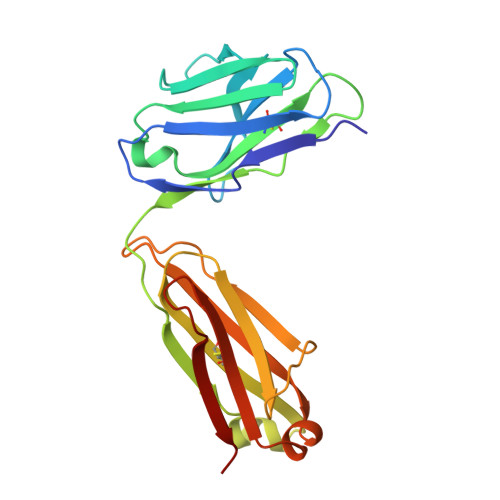
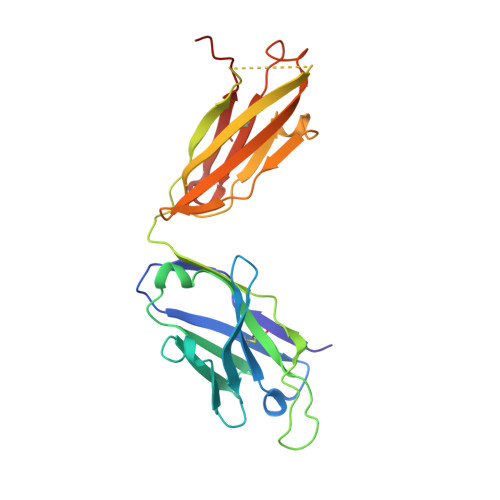
Structure and Molecular Interactions of a Unique Antitumor Antibody Specific for N-Glycolyl GM3.
Krengel, U., Olsson, L.-L., Talavera, A., Rojas, G., Mier, E., Moreno, E.(2004) J Biological Chem 279: 5597-5603
- PubMed: 14627696
- DOI: https://doi.org/10.1074/jbc.M311693200
- Primary Citation of Related Structures:
1RIH - PubMed Abstract:
N-glycolyl GM3 ganglioside is an attractive target antigen for cancer immunotherapy, because this epitope is a molecular marker of certain tumor cells and not expressed in normal human tissues. The murine monoclonal antibody 14F7 specifically recognizes N-glycolyl GM3 and shows no cross-reactivity with the abundant N-acetyl GM3 ganglioside, a close structural homologue of N-glycolyl GM3. Here, we report the crystal structure of the 14F7 Fab fragment at 2.5 A resolution and its molecular model with the saccharide moiety of N-glycolyl GM3, NeuGcalpha3Galbeta4Glcbeta. Fab 14F7 contains a very long CDR H3 loop, which divides the antigen-binding site of this antibody into two subsites. In the docking model, the saccharide ligand is bound to one of these subsites, formed solely by heavy chain residues. The discriminative feature of N-glycolyl GM3 versus N-acetyl GM3, its hydroxymethyl group, is positioned in a hydrophilic cavity, forming hydrogen bonds with the carboxyl group of Asp H52, the indole NH of Trp H33 and the hydroxyl group of Tyr H50. For the hydrophobic methyl group of N-acetyl GM3, this environment would not be favorable, explaining why the antibody specifically recognizes N-glycolyl GM3, but not N-acetyl GM3. Mutation of Asp H52 to hydrophobic residues of similar size completely abolished binding. Our model of the antibodycarbohydrate complex is consistent with binding data for several tested glycolipids as well as for a variety of 14F7 mutants with replaced VL domains.
Organizational Affiliation:
Department of Chemistry and Biosciences, Chalmers University of Technology, P. O. Box 462, SE-40530 Göteborg, Sweden. ute.krengel@molbiotech.chalmers.se