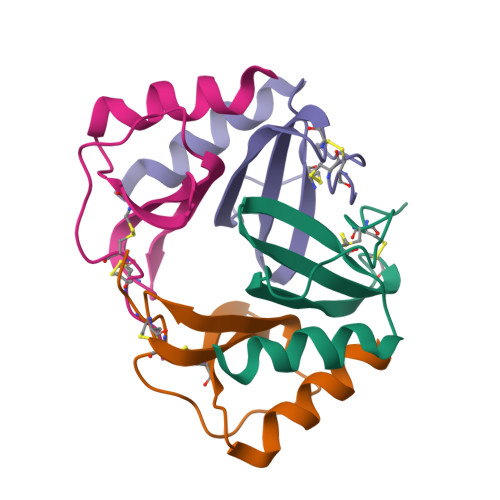
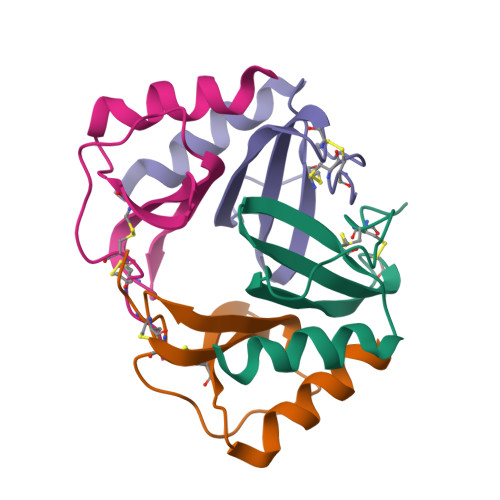
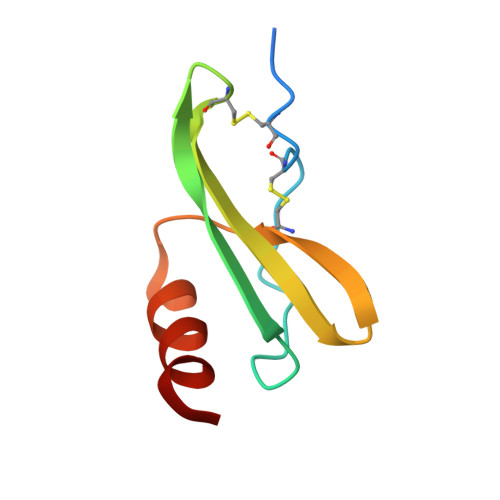
Crystal structure of recombinant human platelet factor 4.
Zhang, X., Chen, L., Bancroft, D.P., Lai, C.K., Maione, T.E.(1994) Biochemistry 33: 8361-8366
- PubMed: 8031770
- DOI: https://doi.org/10.1021/bi00193a025
- Primary Citation of Related Structures:
1RHP - PubMed Abstract:
The crystal structure of human platelet factor 4 (PF4) has been solved to a resolution of 2.4 A by molecular replacement and refined to an R-factor of 24.1%. The structure consists of four polypeptide chains which form a tetrameric unit. N-terminal residues, previously defined as a random coil or extended loop region, form antiparallel beta-sheet-like structures that form noncovalent associations between dimers. These antiparallel beta-sheet-like structures are positioned lateral to the beta-bilayer motif and stabilize the tetrameric unit. A positively charged ring of lysine and arginine side chains encircles the PF4 tetramer sphere, presenting multiple potential sites and orientations for heparin binding. The electrostatic interactions of multiply charged amino acid side chains and hydrogen bonding interactions at the AB/CD dimer interface serve to stabilize the tetrameric structure further.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge 02139.