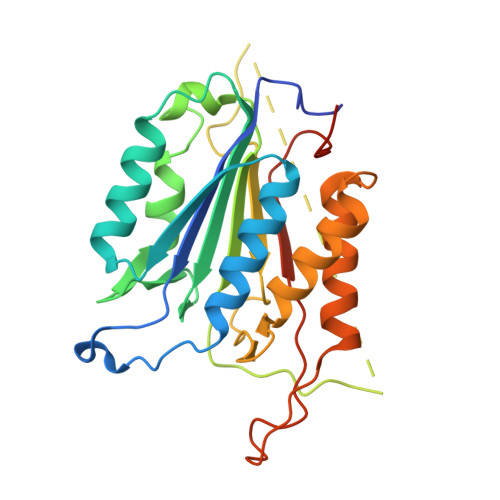
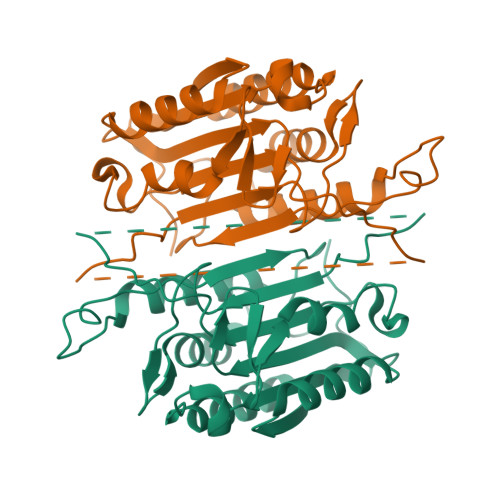
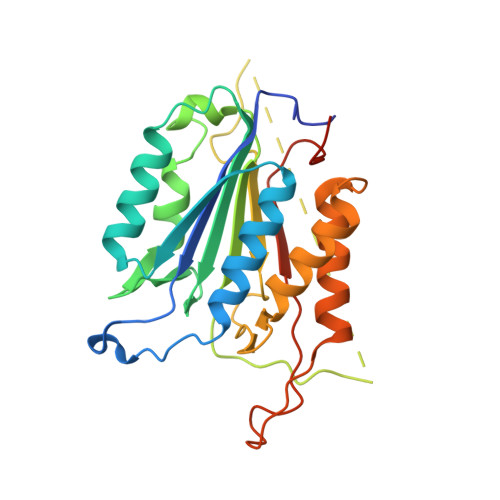
Conformational restrictions in the active site of unliganded human caspase-3
Ni, C.-Z., Li, C., Wu, J.C., Spada, A.P., Ely, K.R.(2003) J Mol Recognit 16: 121-124
- PubMed: 12833566
- DOI: https://doi.org/10.1002/jmr.615
- Primary Citation of Related Structures:
1QX3 - PubMed Abstract:
Caspases are cysteine proteases that play a critical role in the initiation and regulation of apoptosis. These enzymes act in a cascade to promote cell death through proteolytic cleavage of intracellular proteins. Since activation of apoptosis is implicated in human diseases such as cancer and neurodegenerative disorders, caspases are targets for drugs designed to modulate their action. Active caspases are heterodimeric enzymes with two symmetrically arranged active sites at opposite ends of the molecule. A number of crystal structures of caspases with peptides or proteins bound at the active sites have defined the mechanism of action of these enzymes, but molecular information about the active sites before substrate engagement has been lacking. As part of a study of peptidyl inhibitors of caspase-3, we crystallized a complex where the inhibitor did not bind in the active site. Here we present the crystal structure of the unoccupied substrate-binding site of caspase-3. No large conformational differences were apparent when this site was compared with that in enzyme-inhibitor complexes. Instead, the 1.9 A structure reveals critical side chain movements in a hydrophobic pocket in the active site. Notably, the side chain of tyrosine204 is rotated by approximately 90 degrees so that the phenol group occupies the S2 subsite in the active site. Thus, binding of substrate or inhibitors is impeded unless rotation of this side chain opens the area. The positions of these side chains may have important implications for the directed design of inhibitors of caspase-3 or caspase-7.
Organizational Affiliation:
The Burnham Institute, La Jolla, CA 92037, USA.