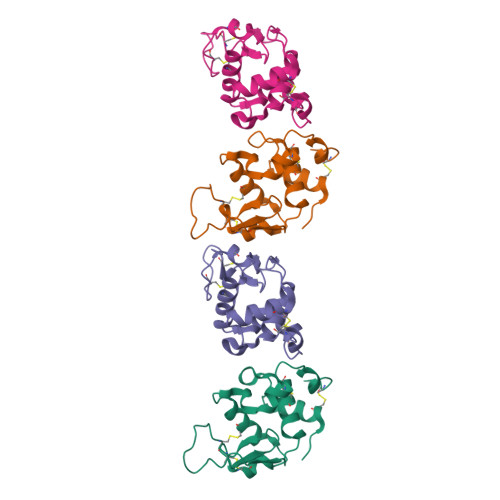
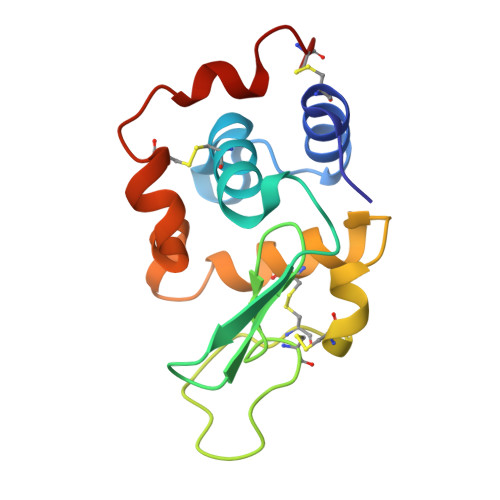
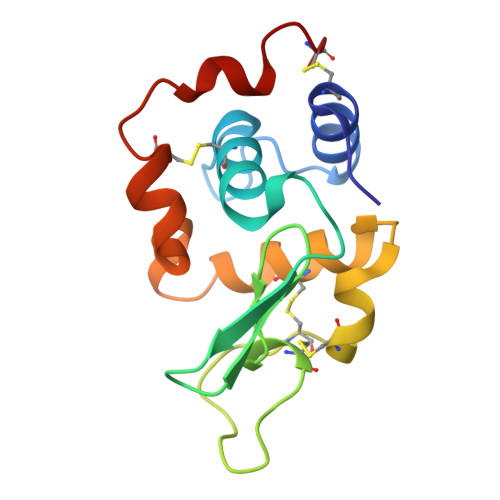
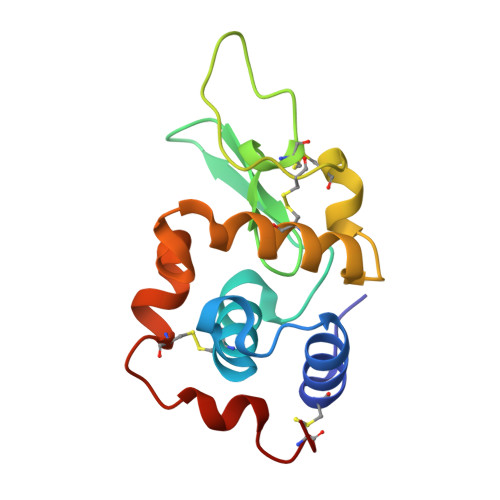
Crystal structure of a mutant human lysozyme with a substituted disulfide bond.
Inaka, K., Kanaya, E., Kikuchi, M., Miki, K.(2001) Proteins 43: 413-419
- PubMed: 11340658
- DOI: https://doi.org/10.1002/prot.1054
- Primary Citation of Related Structures:
1QSW - PubMed Abstract:
The three-dimensional structure of a mutant human lysozyme, W64CC65A, in which a non-native disulfide bond Cys64--Cys81 is substituted for the Cys65--Cys81 of the wild type protein by replacing Trp64 and Cys65 with Cys and Ala, respectively, was determined by X-ray crystallography and refined to an R-value of 0.181, using 33,187 reflections at 1.87-A resolution. The refined model of the W64CC65A protein consisted of four molecules, which were related by two noncrystallographic twofold axes and a translation vector. Although no specific structural differences could be observed among these four molecules, the overall B-factors of each molecule were quite different. The overall structure of W64CC65A, especially in the alpha-helical domain, was found to be quite similar to that of the wild type protein. Moreover, the side-chain conformation of the newly formed Cys64--Cys81 bond was quite similar to that of the Cys65--Cys81 bond of the wild-type protein. However, in the beta-sheet domain, the main-chain atoms of the loop region from positions 66-75 could not be determined, and significant structural changes due to the formation of the non-native disulfide bond could be observed. From these results, it is clear that the loop region of the mutant protein does not fold with the specific folding as observed in the wild-type protein.
Organizational Affiliation:
Research Laboratory of Resources Utilization, Tokyo Institute of Technology, Nagatsuta, Midori-ku, Yokohama, Japan.