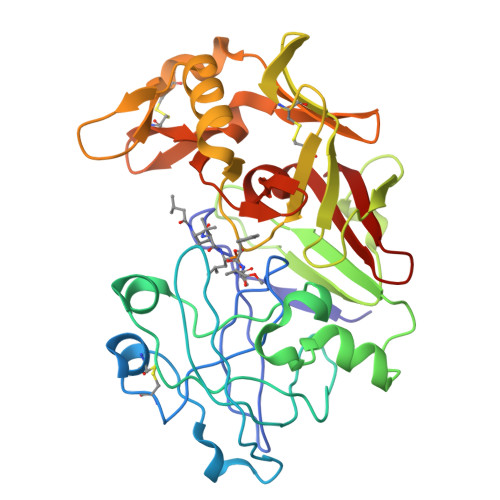
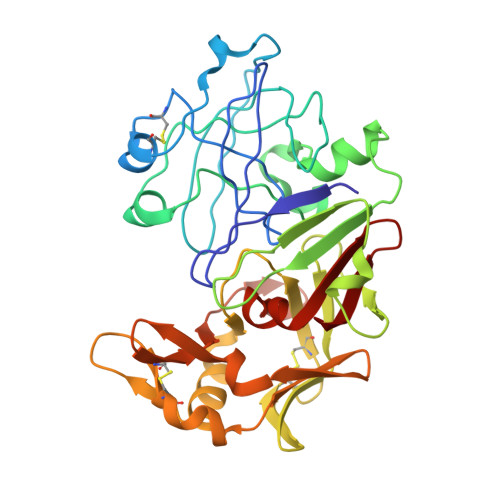
Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Fujinaga, M., Cherney, M.M., Tarasova, N.I., Bartlett, P.A., Hanson, J.E., James, M.N.(2000) Acta Crystallogr D Biol Crystallogr 56: 272-279
- PubMed: 10713513
- DOI: https://doi.org/10.1107/s0907444999016376
- Primary Citation of Related Structures:
1QRP - PubMed Abstract:
The refined crystal structure of the complex between human pepsin and a synthetic phosphonate inhibitor, Iva-Val-Val-Leu(P)-(O)Phe-Ala-Ala-OMe [Iva = isovaleryl, Leu(P) = the phosphinic acid analog of L-leucine, (O)Phe = L-3-phenyllactic acid, OMe = methyl ester], is presented. The structure was refined using diffraction data between 30 and 1.96 A resolution to a final R factor ( summation operator| |F(o)| - |F(c)| | / summation operator|F(o)|, where |F(o)| and |F(c)| are the observed and calculated structure-factor amplitudes, respectively) of 20.0%. The interactions of the inhibitor with the enzyme show the locations of the binding sites on the enzyme from S4 to S3'. Modeling of the inhibitor binding to porcine pepsin shows very similar binding sites, except at S4. Comparison of the complex structure with the structures of related inhibitors bound to penicillopepsin helps to rationalize the observed differences in the binding constants. The convergence of reaction mechanisms and geometries in different families of proteinases is also discussed.
Organizational Affiliation:
Medical Research Council of Canada Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.