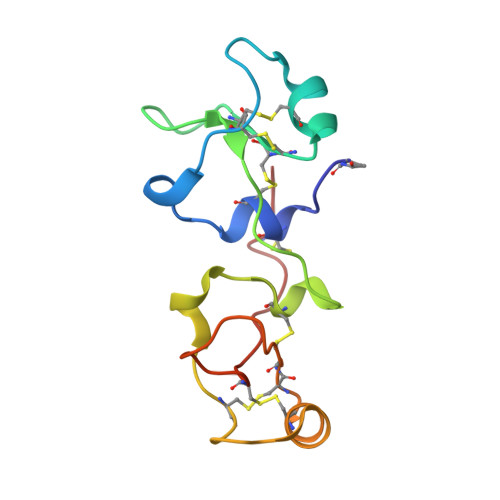
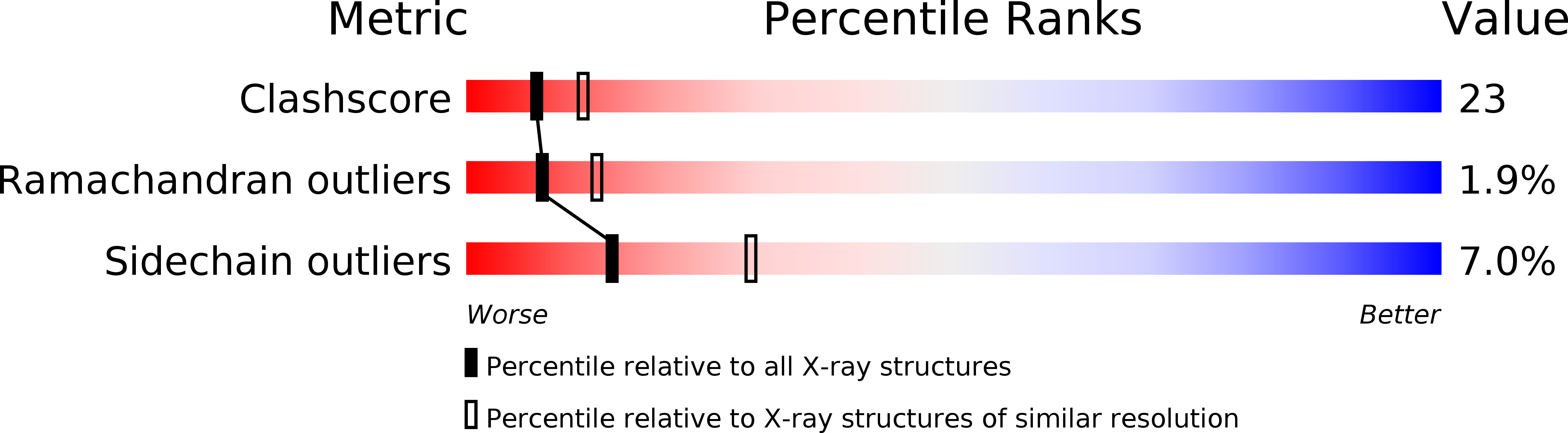Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Gajhede, M., Petersen, T.N., Henriksen, A., Petersen, J.F., Dauter, Z., Wilson, K.S., Thim, L.(1993) Structure 1: 253-262
- PubMed: 8081739
- DOI: https://doi.org/10.1016/0969-2126(93)90014-8
- Primary Citation of Related Structures:
1PSP - PubMed Abstract:
The trefoil peptides are a rapidly growing family of peptides, mainly found in the gastrointestinal tract. There is circumstantial evidence that they stabilize the mucus layer, and may affect the rate of healing of the mucosal epithelium. We have determined the structure of porcine pancreatic spasmolytic polypeptide (PSP) to 2.5 A resolution. The polypeptide contains two trefoil domains. The domain structure is compact, and is composed of a central short antiparallel beta-sheet with one short helix above and one below it. This is a novel motif. The two domains are related by two-fold symmetry, and each domain contains a cleft. The cleft within each domain could accommodate a polysaccharide chain, and may therefore be responsible for binding mucin glycoproteins. We suggest that PSP may cross-link glycoproteins, explaining its ability to stabilize the mucus layer.
Organizational Affiliation:
Department of Chemistry, H.C. Orsted Institute, University of Copenhagen, Denmark.