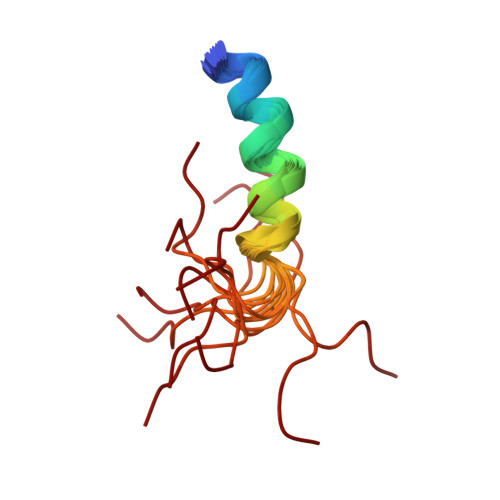
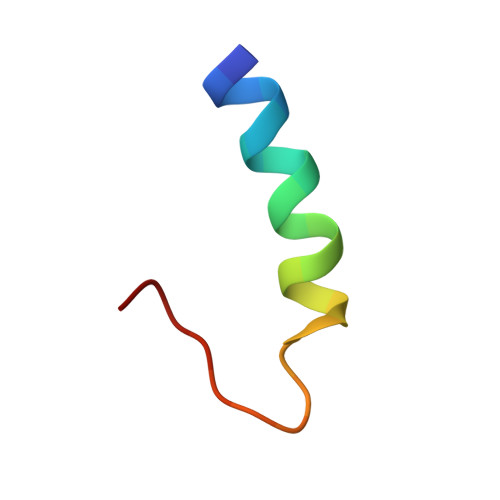
Solution structure of the cytoplasmic domain of phopholamban: phosphorylation leads to a local perturbation in secondary structure.
Mortishire-Smith, R.J., Pitzenberger, S.M., Burke, C.J., Middaugh, C.R., Garsky, V.M., Johnson, R.G.(1995) Biochemistry 34: 7603-7613
- PubMed: 7779806
- DOI: https://doi.org/10.1021/bi00023a006
- Primary Citation of Related Structures:
1PLP - PubMed Abstract:
Peptides representing the N-terminal domain (Ia) of the cardiac sarcoplasmic reticulum protein phospholamban (residues 1-25 [PLB(1-25)] and a phosphorylated form [pPLB(1-25)]) were synthesized and their conformations examined using circular dichroism and nuclear magnetic resonance spectroscopy. In aqueous solution, both PLB(1-25) and pPLB(1-25) adopt a primarily disordered conformation. In 30% trifluoroethanol/10 mM phosphate, PLB(1-25) exhibits a CD spectrum consistent with 60% helical structure. This value decreases to 27% for the phosphorylated peptide. CD spectra in 2% SDS indicate 40% alpha-helix for PLB(1-25) and 20% for pPLB(1-25). Full chemical shift assignments were obtained by conventional homonuclear NMR methodologies for both PLB(1-25) and pPLB(1-25) in 30% trifluoroethanol/water and 300 mM SDS. The solution structure of PLB(1-25) in 30% TFE/water was determined from distance geometry calculations using 54 NOE distance constraints and 17 torsion angle constraints. In the family of 20 calculated conformers, the root mean square deviation from the mean structure is 0.79 A for backbone heavy atoms of residues 1-17. The structure comprises a regular alpha-helix extending from M1 to S16 with the remaining C-terminal residues disordered. The calculated structure is supported by analysis of C alpha H secondary shifts which are significantly negative for residues 1-16. Chemical shift degeneracy is substantially more extensive in the phospho form and precludes a direct comparison of calculated structures. However, the magnitudes of upfield secondary shifts are decreased by 20% in residues 1-11 and are not significantly helical for residues 12-16 according to the criteria of Wishart et al. [(1992) Biochemistry 31, 1647-1651]. 3JHN alpha coupling constants measured for I12, R13, A15, and S16 also suggest that residues 12-16 undergo a local unwinding of the helix upon phosphorylation. Similar results are obtained for PLB(1-25) and pPLB(1-25) in 300 mM perdeuterated sodium dodecyl sulfate except that differences in backbone dynamics for the helical and nonhelical regions of the peptide are evident in the DQF-COSY line shapes for fingerprint cross-peaks. This disruption of structure at the C-terminus of the helix suggests a model for phosphorylation-induced dissociation of the PLB/Ca(2+)-ATPase complex.
Organizational Affiliation:
Neuroscience Research Centre, Merck Sharp and Dohme Research Laboratories, Harlow, Essex, United Kingdom.