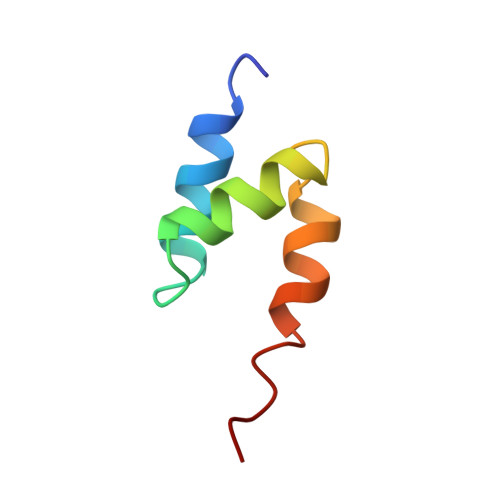
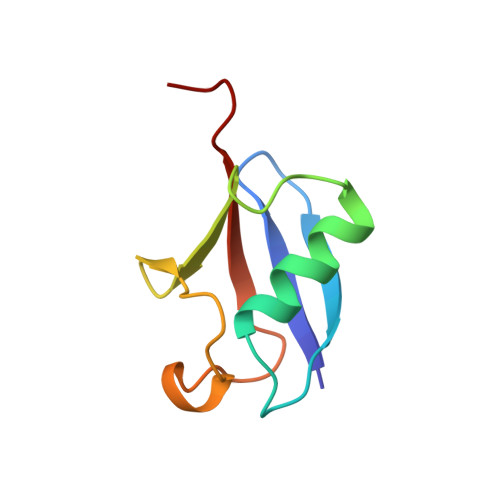
Solution Structure of a CUE-Ubiquitin Complex Reveals a Conserved Mode of Ubiquitin Binding
Kang, R.S., Daniels, C.M., Francis, S.A., Shih, S.C., Salerno, W.J., Hicke, L., Radhakrishnan, I.(2003) Cell 113: 621-630
- PubMed: 12787503
- DOI: https://doi.org/10.1016/s0092-8674(03)00362-3
- Primary Citation of Related Structures:
1OTR - PubMed Abstract:
Monoubiquitination serves as a regulatory signal in a variety of cellular processes. Monoubiquitin signals are transmitted by binding to a small but rapidly expanding class of ubiquitin binding motifs. Several of these motifs, including the CUE domain, also promote intramolecular monoubiquitination. The solution structure of a CUE domain of the yeast Cue2 protein in complex with ubiquitin reveals intermolecular interactions involving conserved hydrophobic surfaces, including the Leu8-Ile44-Val70 patch on ubiquitin. The contact surface extends beyond this patch and encompasses Lys48, a site of polyubiquitin chain formation. This suggests an occlusion mechanism for inhibiting polyubiquitin chain formation during monoubiquitin signaling. The CUE domain shares a similar overall architecture with the UBA domain, which also contains a conserved hydrophobic patch. Comparative modeling suggests that the UBA domain interacts analogously with ubiquitin. The structure of the CUE-ubiquitin complex may thus serve as a paradigm for ubiquitin recognition and signaling by ubiquitin binding proteins.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Cell Biology, Northwestern University, Evanston, IL 60208, USA.