BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Gordon, N.C., Pan, B., Hymowitz, S.G., Yin, J.P., Kelley, R.F., Cochran, A.G., Yan, M., Dixit, V.M., Fairbrother, W.J., Starovasnik, M.A.(2003) Biochemistry 42: 5977-5983
- PubMed: 12755599
- DOI: https://doi.org/10.1021/bi034017g
- Primary Citation of Related Structures:
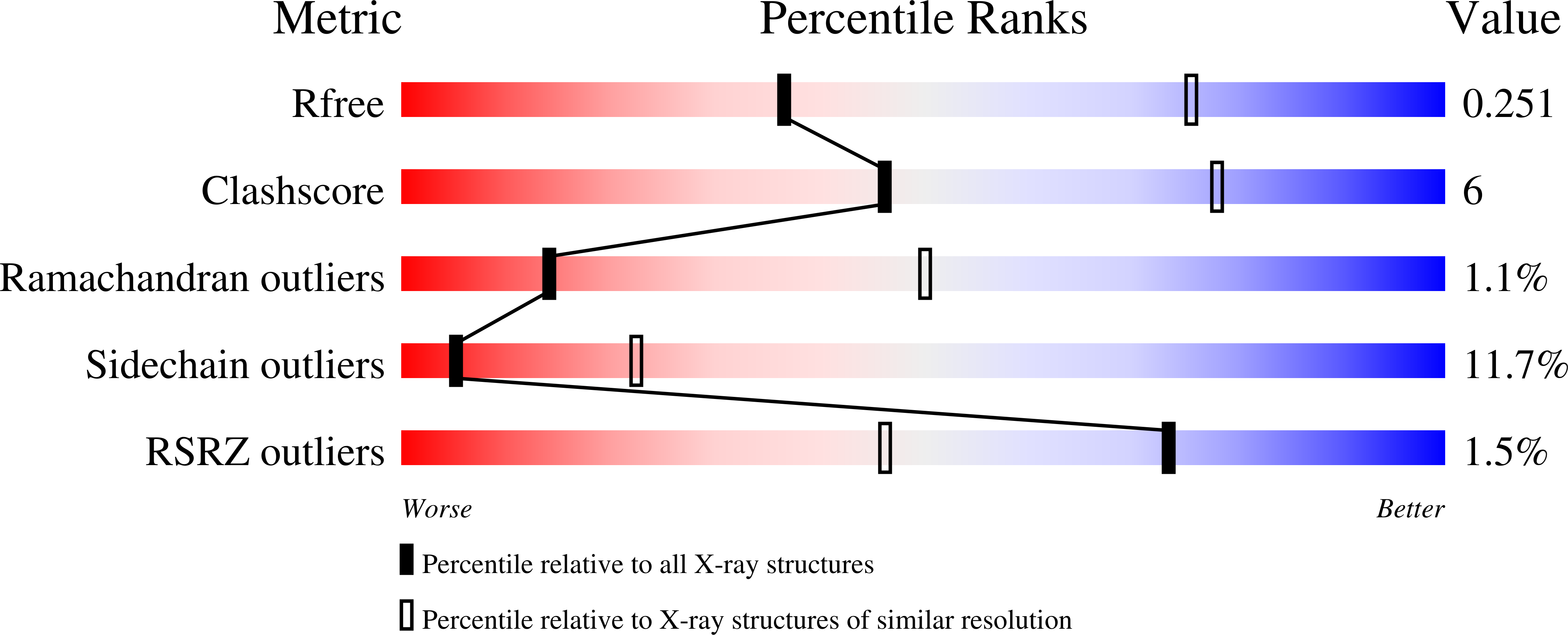
1OSG, 1OSX - PubMed Abstract:
BAFF/BLyS, a member of the tumor necrosis family (TNF) superfamily of ligands, is a crucial survival factor for B cells. BAFF binds three receptors, TACI, BCMA, and BR3, with signaling through BR3 being essential for promoting B cell function. Typical TNF receptor (TNFR) family members bind their cognate ligands through interactions with two cysteine-rich domains (CRDs). However, the extracellular domain (ECD) of BR3 consists of only a partial CRD, with cysteine spacing distinct from other modules described previously. Herein, we report the solution structure of the BR3 ECD. A core region of only 19 residues adopts a stable structure in solution. The BR3 fold is analogous to the first half of a canonical TNFR CRD but is stabilized by an additional noncanonical disulfide bond. BAFF-binding determinants were identified by shotgun alanine-scanning mutagenesis of the BR3 ECD expressed on phage. Several of the key BAFF-binding residues are presented from a beta-turn that we have shown previously to be sufficient for ligand binding when transferred to a structured beta-hairpin scaffold [Kayagaki, N., Yan, M., Seshasayee, D., Wang, H., Lee, W., French, D. M., Grewal, I. S., Cochran, A. G., Gordon, N. C., Yin, J., Starovasnik, M. A, and Dixit, V. M. (2002) Immunity 10, 515-524]. Outside of the turn, mutagenesis identifies additional hydrophobic contacts that enhance the BAFF-BR3 interaction. The crystal structure of the minimal hairpin peptide, bhpBR3, in complex with BAFF reveals intimate packing of the six-residue BR3 turn into a cavity on the ligand surface. Thus, BR3 binds BAFF through a highly focused interaction site, unprecedented in the TNFR family.
Organizational Affiliation:
Department of Protein Engineering, Genentech, Inc., One DNA Way, South San Francisco, California 94080, USA.