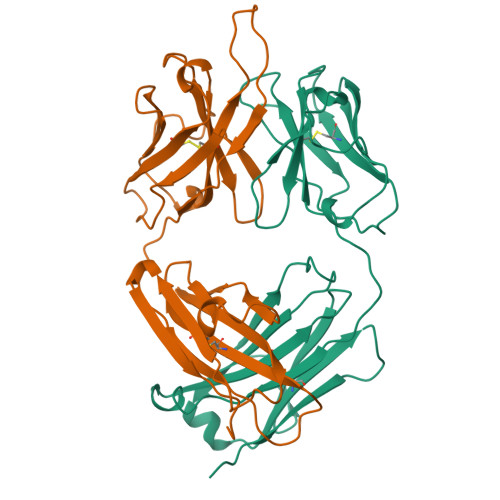
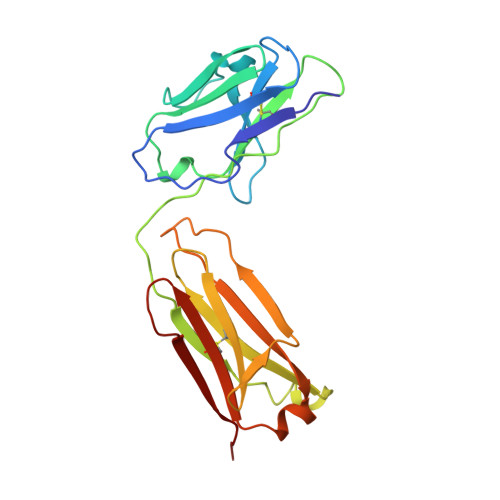
Crystal structure of the OPG2 Fab. An antireceptor antibody that mimics an RGD cell adhesion site.
Kodandapani, R., Veerapandian, B., Kunicki, T.J., Ely, K.R.(1995) J Biological Chem 270: 2268-2273
- PubMed: 7836460
- DOI: https://doi.org/10.1074/jbc.270.5.2268
- Primary Citation of Related Structures:
1OPG - PubMed Abstract:
Cell surface receptors called integrins mediate diverse cell adhesion phenomena through recognition of the sequence arginine-glycine-aspartic acid (RGD) present in proteins such as fibronectin and fibrinogen. Platelet aggregation in hemostasis is mediated by the binding of fibrinogen to the gpIIb/IIIa integrin. The OPG2 antibody binds the gpIIb/IIIa receptor and acts as a ligand mimic due to the presence of an arginine-tyrosine-aspartic acid (RYD) sequence in the CDR3 loop of the heavy chain. The RYD loop and side chains are ordered in the 2.0-A resolution crystal structure of the Fab fragment from this antireceptor antibody. Moreover, the RYD loop assumes two clearly defined conformations that may correspond to the orientations of the loop in the free state or bound to integrin. This molecule will serve as a tool for understanding protein-integrin recognition in platelet aggregation and other RGD-mediated cell adhesion interactions.
Organizational Affiliation:
Cancer Research Center, La Jolla Cancer Research Foundation, California 92037.