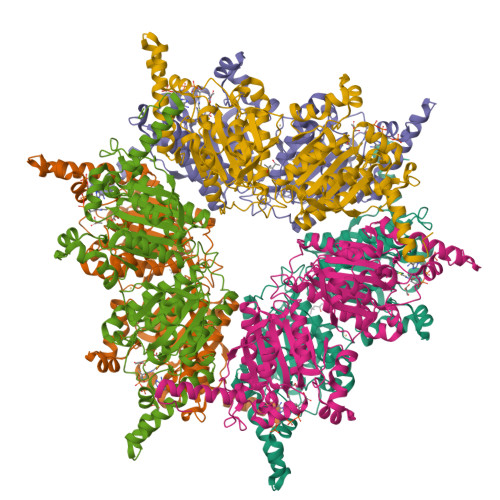
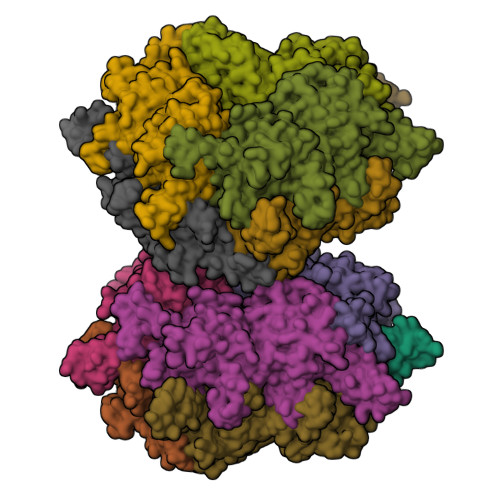
Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Hall, P.R., Wang, Y.-F., Rivera-Hainaj, R.E., Zheng, X., Pustai-Carey, M., Carey, P.R., Yee, V.C.(2003) EMBO J 22: 2334-2347
- PubMed: 12743028
- DOI: https://doi.org/10.1093/emboj/cdg244
- Primary Citation of Related Structures:
1ON3, 1ON9 - PubMed Abstract:
Transcarboxylase from Propionibacterium shermanii is a 1.2 MDa multienzyme complex that couples two carboxylation reactions, transferring CO(2)(-) from methylmalonyl-CoA to pyruvate, yielding propionyl-CoA and oxaloacetate. The 1.9 A resolution crystal structure of the central 12S hexameric core, which catalyzes the first carboxylation reaction, has been solved bound to its substrate methylmalonyl-CoA. Overall, the structure reveals two stacked trimers related by 2-fold symmetry, and a domain duplication in the monomer. In the active site, the labile carboxylate group of methylmalonyl-CoA is stabilized by interaction with the N-termini of two alpha-helices. The 12S domains are structurally similar to the crotonase/isomerase superfamily, although only domain 1 of each 12S monomer binds ligand. The 12S reaction is similar to that of human propionyl-CoA carboxylase, whose beta-subunit has 50% sequence identity with 12S. A homology model of the propionyl-CoA carboxylase beta-subunit, based on this 12S crystal structure, provides new insight into the propionyl-CoA carboxylase mechanism, its oligomeric structure and the molecular basis of mutations responsible for enzyme deficiency in propionic acidemia.
Organizational Affiliation:
Department of Molecular Cardiology and Center for Structural Biology, Lerner Research Institute, Cleveland Clinic Foundation, OH 44195, USA.