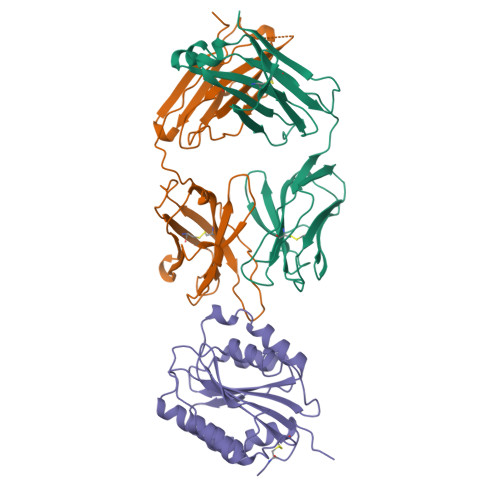
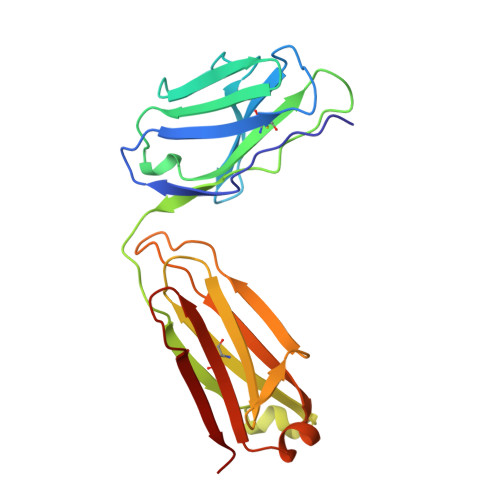
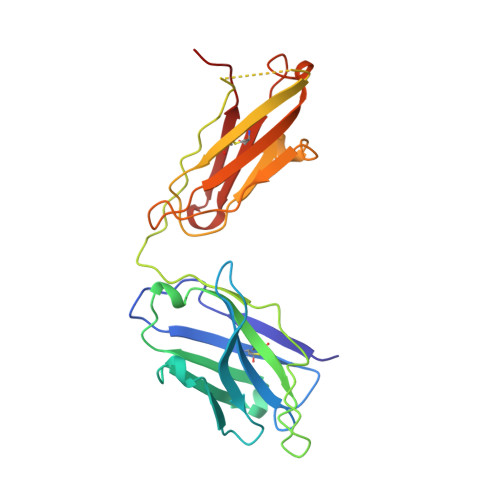
Crystal structure of the von Willebrand factor A1 domain in complex with the function blocking NMC-4 Fab.
Celikel, R., Varughese, K.I., Madhusudan, Yoshioka, A., Ware, J., Ruggeri, Z.M.(1998) Nat Struct Biol 5: 189-194
- PubMed: 9501911
- DOI: https://doi.org/10.1038/nsb0398-189
- Primary Citation of Related Structures:
1OAK - PubMed Abstract:
The presence of one or more copies of von Willebrand factor type A domains identifies a superfamily of proteins usually involved in biological processes controlled by specific molecular interactions, often adhesive in nature. We have solved the crystal structure of the prototypic von Willebrand factor A1 domain, essential for the antihemorrhagic activity of platelets, in complex with the function blocking antibody, NMC-4, at 2.2 A resolution. This has led to the recognition of a putative binding groove for the platelet receptor, glycoprotein Ib alpha, formed by two adjacent alpha-helices and a beta-strand. The structure also shows a contact interface between A1 domain pairs, suggesting a hypothetical mechanism for the regulation of protein assembly and heterologous ligand binding mediated by homophilic interactions of type A domains.
Organizational Affiliation:
Roon Research Center for Arteriosclerosis and Thrombosis, The Scripps Research Institute, La Jolla, California 92037, USA.