Solution structure of the cyclic peptide contryphan-Vn, a Ca2+-dependent K+ channel modulator
Eliseo, T., Cicero, D.O., Romeo, C., Schinina, M.E., Massilia, G.R., Polticelli, F., Ascenzi, P., Paci, M.(2004) Biopolymers 74: 189-198
- PubMed: 15150794
- DOI: https://doi.org/10.1002/bip.20025
- Primary Citation of Related Structures:
1NXN - PubMed Abstract:
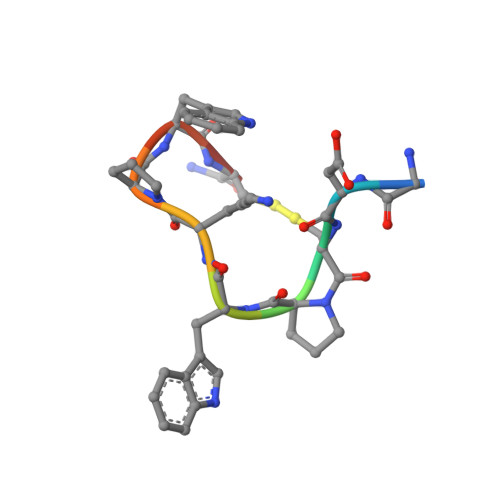
The solution structure of contryphan-Vn, a cyclic peptide with a double cysteine S-S bridge and containing a D-tryptophan extracted from the venom of the cone snail Conus ventricosus, has been determined by NMR spectroscopy using a variety of homonuclear and heteronuclear NMR methods and restrained molecular dynamics simulations. The main conformational features of backbone contryphan-Vn are a type IV beta-turn from Gly 1 to Lys 6 and a type I beta-turn from Lys 6 to Cys 9. As already found in other contryphans, one of the two prolines--the Pro4--is mainly in the cis conformation while Pro7 is trans. A small hydrophobic region probably partly shielded from solvent constituted from the close proximity of side chains of Pro7 and Trp8 was observed together with a persistent salt bridge between Asp2 and Lys6, which has been revealed by the diagnostic observation of specific nuclear Overhauser effects. The salt bridge was used as a restraint in the molecular dynamics in vacuum but without inserting explicit electrostatic contribution in the calculations. The backbone of the unique conformational family found of contryphan-Vn superimposes well with those of contryphan-Sm and contryphan-R. This result indicates that the contryphan structural motif represents a robust and conserved molecular scaffold whose main structural determinants are the size of the intercysteine loop and the presence and location in the sequence of the D-Trp and the two Pro residues.
Organizational Affiliation:
Department of Chemistry, University of Rome La Sapienza, Piazzale Aldo Moro 5, 00198 Rome, Italy.