An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
Bartolucci, S., De Simone, G., Galdiero, S., Improta, R., Menchise, V., Pedone, C., Pedone, E., Saviano, M.(2003) J Bacteriol 185: 4285-4289
- PubMed: 12837806
- DOI: https://doi.org/10.1128/JB.185.14.4285-4289.2003
- Primary Citation of Related Structures:
1NSW, 1NW2 - PubMed Abstract:
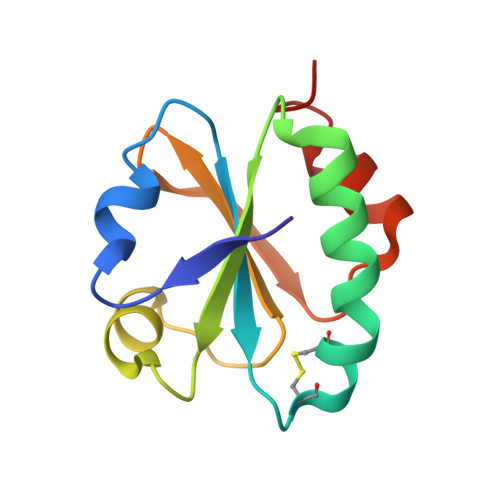
We report a crystallographic and computational analysis of two mutant forms of the Alicyclobacillus acidocaldarius thioredoxin (BacTrx) done in order to evaluate the contribution of two specific amino acids to the thermostability of BacTrx. Our results suggest that the thermostability of BacTrx may be modulated by mutations affecting the overall electrostatic energy of the protein.
Organizational Affiliation:
Dipartimento di Chimica Biologica, University of Naples Federico II, Istituto di Biostrutture e Bioimmagini-CNR, 80134 Naples, Italy.