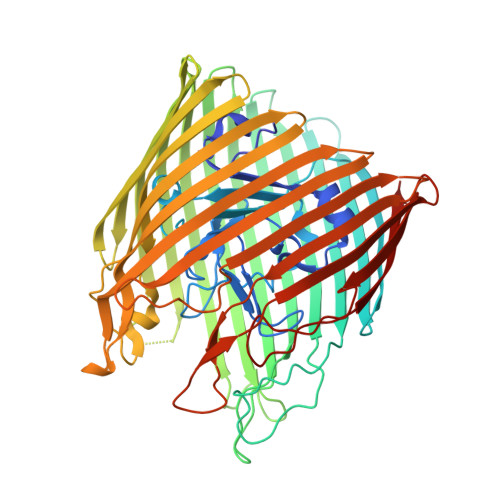
Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
CHIMENTO, D.P., MOHANTY, A.K., KADNER, R.J., WIENER, M.C.(2003) Nat Struct Biol 10: 394-401
- PubMed: 12652322
- DOI: https://doi.org/10.1038/nsb914
- Primary Citation of Related Structures:
1NQE, 1NQF, 1NQG, 1NQH - PubMed Abstract:
The outer membranes of Gram-negative bacteria possess transport proteins essential for uptake of scarce nutrients. In TonB-dependent transporters, a conserved sequence of seven residues, the Ton box, faces the periplasm and interacts with the inner membrane TonB protein to energize an active transport cycle. A critical mechanistic step is the structural change in the Ton box of the transporter upon substrate binding; this essential transmembrane signaling event increases the affinity of the transporter for TonB and enables active transport to proceed. We have solved crystal structures of BtuB, the outer membrane cobalamin transporter from Escherichia coli, in the absence and presence of cyanocobalamin (vitamin B(12)). In these structures, the Ton box is ordered and undergoes a conformational change in the presence of bound substrate. Calcium has been implicated as a necessary factor for the high-affinity binding (K(d) approximately 0.3 nM) of cyanocobalamin to BtuB. We observe two bound calcium ions that order three extracellular loops of BtuB, thus providing a direct (and unusual) structural role for calcium.
Organizational Affiliation:
Department of Microbiology, University of Virginia, Charlottesville, Virginia 22908, USA.