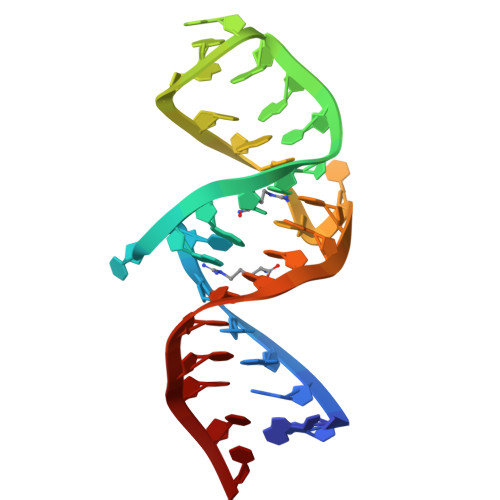
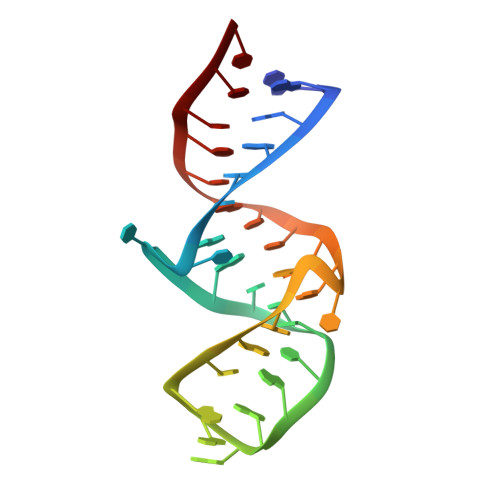
Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Matsugami, A., Kobayashi, S., Ouhashi, K., Uesugi, S., Yamamoto, R., Taira, K., Nishikawa, S., Kumar, P.K.R., Katahira, M.(2003) Structure 11: 533-545
- PubMed: 12737819
- DOI: https://doi.org/10.1016/s0969-2126(03)00069-8
- Primary Citation of Related Structures:
1NBK - PubMed Abstract:
An RNA aptamer containing two binding sites exhibits extremely high affinity to the HIV Tat protein. We have determined the structure of the aptamer complexed with two argininamide molecules. Two adjacent U:A:U base triples were formed, which widens the major groove to make space for the two argininamide molecules. The argininamide molecules bind to the G bases through hydrogen bonds. The binding is stabilized through stacking interactions. The structure of the aptamer complexed with a Tat-derived arginine-rich peptide was also characterized. It was suggested that the aptamer structure is similar for both complexes and that the aptamer interacts with two different arginine residues of the peptide simultaneously at the two binding sites, which could explain the high affinity to Tat.
Organizational Affiliation:
Department of Environment and Natural Sciences, Graduate School of Environment and Information Sciences, Yokohama National University, 79-7 Tokiwadai, Hodogaya-ku, 240-8501, Yokohama, Japan.