Solution structure of the human Grb7-SH2 domain/erbB2 peptide complex and structural basis for Grb7 binding to ErbB2
Ivancic, M., Daly, R.J., Lyons, B.A.(2003) J Biomol NMR 27: 205-219
- PubMed: 12975581
- DOI: https://doi.org/10.1023/a:1025498409113
- Primary Citation of Related Structures:
1MW4 - PubMed Abstract:
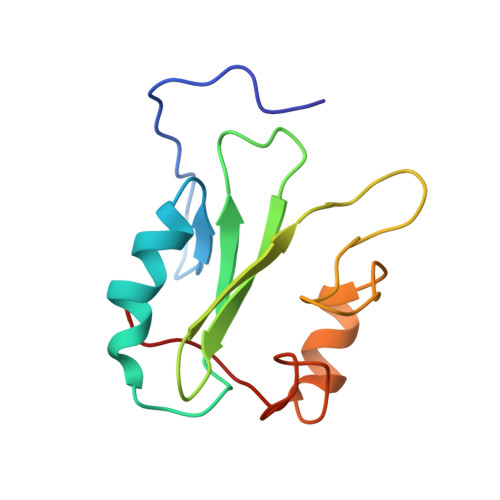
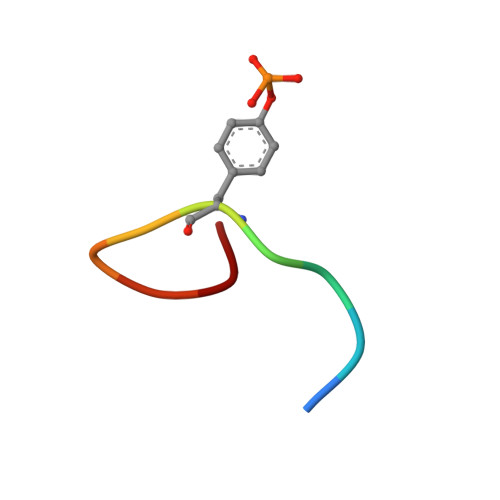
The solution structure of the hGrb7-SH2 domain in complex with a ten amino acid phosphorylated peptide ligand representative of the erbB2 receptor tyrosine kinase (pY1139) is presented as determined by nuclear magnetic resonance methods. The hGrb7-SH2 domain structure reveals the Src homology 2 domain topology consisting of a central beta-sheet capped at each end by an alpha-helix. The presence of a four residue insertion in the region between beta-strand E and the EF loop and resulting influences on the SH2 domain/peptide complex structure are discussed. The binding conformation of the erbB2 peptide is in a beta-turn similar to that found in phosphorylated tyrosine peptides bound to the Grb2-SH2 domain. To our knowledge this is only the second example of an SH2 domain binding its naturally occurring ligands in a turn, instead of extended, conformation. Close contacts between residues responsible for binding specificity in hGrb7-SH2 and the erbB2 peptide are characterized and the potential effect of mutation of these residues on the hGrb7-SH2 domain structure is discussed.
Organizational Affiliation:
Department of Biochemistry, University of Vermont College of Medicine Burlington, VT 05405, U.S.A.