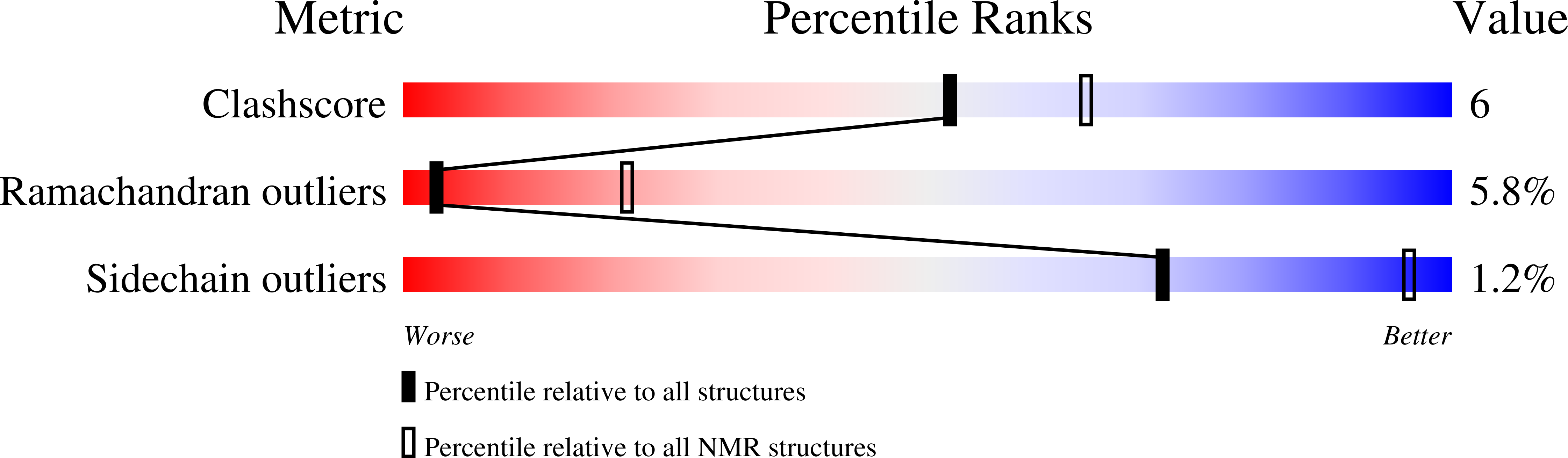Solution structure of termicin, an antimicrobial peptide from the termite Pseudacanthotermes spiniger
Da Silva, P., Jouvensal, L., Lamberty, M., Bulet, P., Caille, A., Vovelle, F.(2003) Protein Sci 12: 438-446
- PubMed: 12592014
- DOI: https://doi.org/10.1110/ps.0228303
- Primary Citation of Related Structures:
1MM0 - PubMed Abstract:
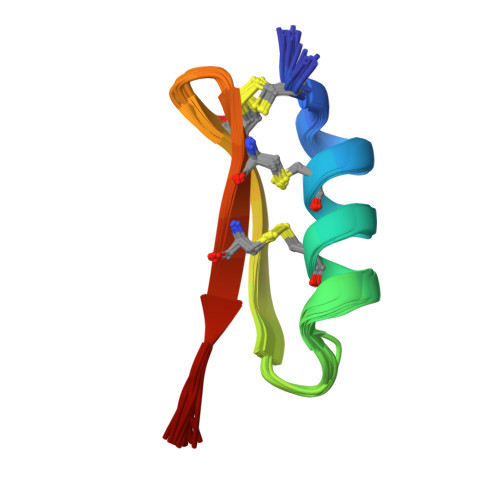
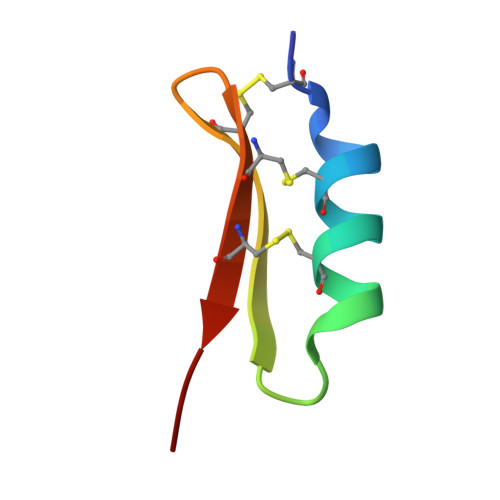
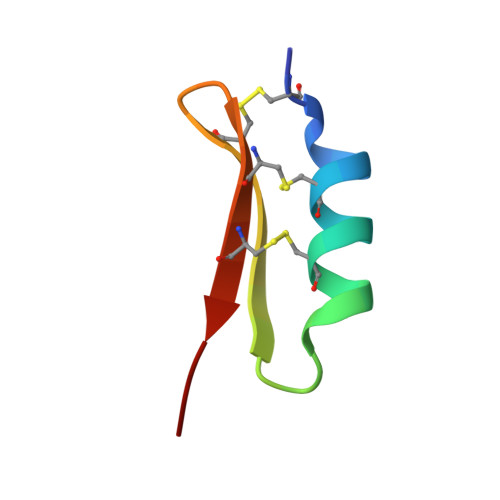
The solution structure of termicin from hemocytes of the termite Pseudacanthotermes spiniger was determined by proton two-dimensional nuclear magnetic resonance spectroscopy and molecular modeling techniques. Termicin is a cysteine-rich antifungal peptide also exhibiting a weak antibacterial activity. The global fold of termicin consists of an alpha-helical segment (Phe4-Gln14) and a two-stranded (Phe19-Asp25 and Gln28-Phe33) antiparallel beta-sheet forming a "cysteine stabilized alphabeta motif" (CSalphabeta) also found in antibacterial and antifungal defensins from insects and from plants. Interestingly, termicin shares more structural similarities with the antibacterial insect defensins and with MGD-1, a mussel defensin, than with the insect antifungal defensins such as drosomycin and heliomicin. These structural comparisons suggest that global fold alone does not explain the difference between antifungals and antibacterials. The antifungal properties of termicin may be related to its marked hydrophobicity and its amphipatic structure as compared to the antibacterial defensins. [SWISS-PROT accession number: Termicin (P82321); PDB accession number: 1MM0.]
Organizational Affiliation:
Centre de Biophysique Moléculaire, UPR 4301 CNRS affiliated at Orléans University, 45071 Orléans cedex 2, France.