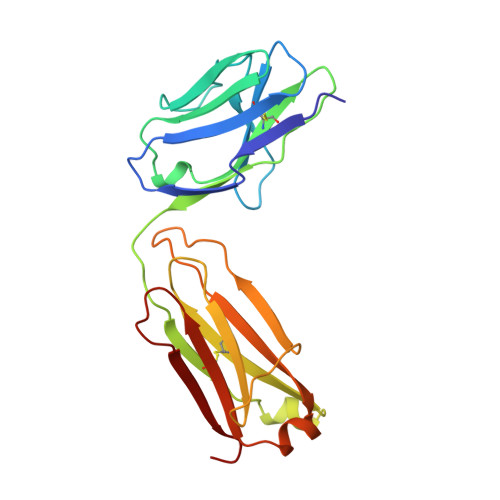
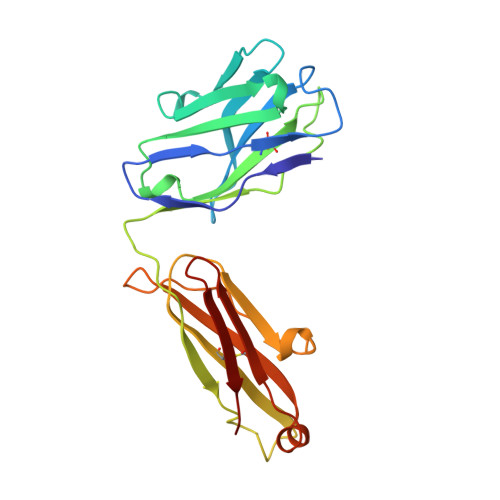
Structure of the fab fragment of SDZ CHI621: a chimeric antibody against CD25.
Mikol, V.(1996) Acta Crystallogr D Biol Crystallogr 52: 534-542
- PubMed: 15299676
- DOI: https://doi.org/10.1107/S0907444996000704
- Primary Citation of Related Structures:
1MIM - PubMed Abstract:
A specific drug targeted to the IL-2 receptor on activated T lymphocytes could limit acute immunological rejection during organ transplantation. A high-affinity monoclonal antibody directed against the alpha-chain of the IL-2 receptor (CD25) was chimerized with the constant regions of the human IgG1 heavy and k light chain resulting in SDZ CHII621 [Amlot, Rawlings, Fernando, Griffin, Heinrich, Schreier, Castaigne, Moore & Sweny (1995). Transplantation, 60, 748-756]. The Fab fragment of SDZ CHI621 has been purified and crystallized (P2(l), a = 39.58, b = 59.76, c = 102.09 A, beta = 99.98 degrees ). Its structure has been determined by molecular replacement and refined at 2.6 A to a crystallographic R factor of 19.7%. The protein exhibits the typical immunoglobulin fold. The complementary determining regions (CDR's) 1 and 2 of both heavy and light chains show similar conformations to other known reported structures whereas the CDR3 from the light chain seems to adopt a novel type of conformation. There is a network of interactions which maintain the CDR3 of both chains together and limit their solvent accessibility. The interaction between V(L) and C(L) has been strengthened by the chimerization whereas that between V(H) and C(H)1 has been weakened.
Organizational Affiliation:
Preclinical Research, Sandoz Pharma AG, Basel, Switzerland. mikol@rp.fr