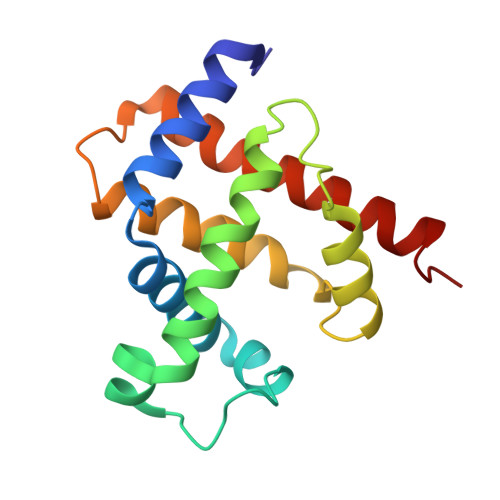
X-ray crystallographic studies of seal myoglobin. The molecule at 2.5 A resolution.
Scouloudi, H., Baker, E.N.(1978) J Mol Biol 126: 637-660
- PubMed: 745243
- DOI: https://doi.org/10.1016/0022-2836(78)90013-x
- Primary Citation of Related Structures:
1MBS