Function and solution structure of huwentoxin-IV, a potent neuronal tetrodotoxin (TTX)-sensitive sodium channel antagonist from Chinese bird spider Selenocosmia huwena
Peng, K., Shu, Q., Liu, Z., Liang, S.P.(2002) J Biol Chem 277: 47564-47571
- PubMed: 12228241
- DOI: https://doi.org/10.1074/jbc.M204063200
- Primary Citation of Related Structures:
1MB6 - PubMed Abstract:
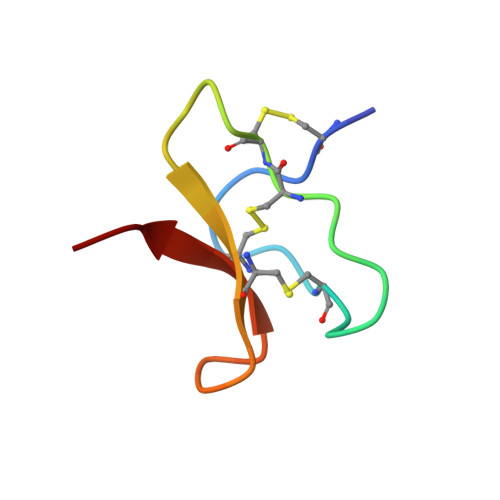
We have isolated a highly potent neurotoxin from the venom of the Chinese bird spider, Selenocosmia huwena. This 4.1-kDa toxin, which has been named huwentoxin-IV, contains 35 residues with three disulfide bridges: Cys-2-Cys-17, Cys-9-Cys-24, and Cys-16-Cys-31, assigned by a chemical strategy including partial reduction of the toxin and sequence analysis of the modified intermediates. It specifically inhibits the neuronal tetrodotoxin-sensitive (TTX-S) voltage-gated sodium channel with the IC(50) value of 30 nm in adult rat dorsal root ganglion neurons, while having no significant effect on the tetrodotoxin-resistant (TTX-R) voltage-gated sodium channel. This toxin seems to be a site I toxin affecting the sodium channel through a mechanism quite similar to that of TTX: it suppresses the peak sodium current without altering the activation or inactivation kinetics. The three-dimensional structure of huwentoxin-IV has been determined by two-dimensional (1)H NMR combined with distant geometry and simulated annealing calculation by using 527 nuclear Overhauser effect constraints and 14 dihedral constraints. The resulting structure is composed of a double-stranded antiparallel beta-sheet (Leu-22-Ser-25 and Trp-30-Tyr-33) and four turns (Glu-4-Lys-7, Pro-11-Asp-14, Lys-18-Lys-21 and Arg-26-Arg-29) and belongs to the inhibitor cystine knot structural family. After comparison with other toxins purified from the same species, we are convinced that the positively charged residues of loop IV (residues 25-29), especially residue Arg-26, must be crucial to its binding to the neuronal tetrodotoxin-sensitive voltage-gated sodium channel.
Organizational Affiliation:
College of Life Sciences, Hunan Normal University, Changsha 410081, People's Republic of China.