Transition state stabilization by a catalytic RNA
Rupert, P.B., Massey, A.P., Sigurdsson, S.T., Ferre-D'Amare, A.R.(2002) Science 298: 1421-1424
- PubMed: 11298439
- DOI: https://doi.org/10.1038/35071009
- Primary Citation of Related Structures:
1M5K - PubMed Abstract:
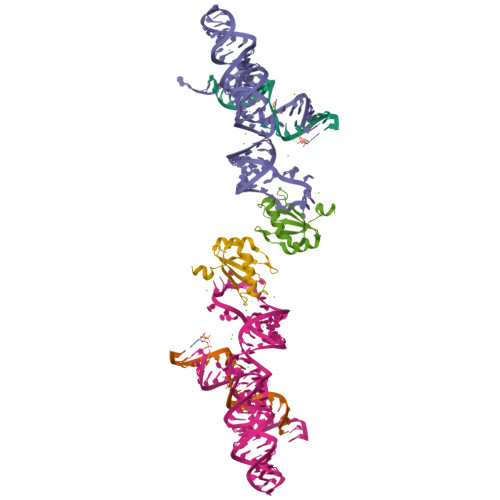
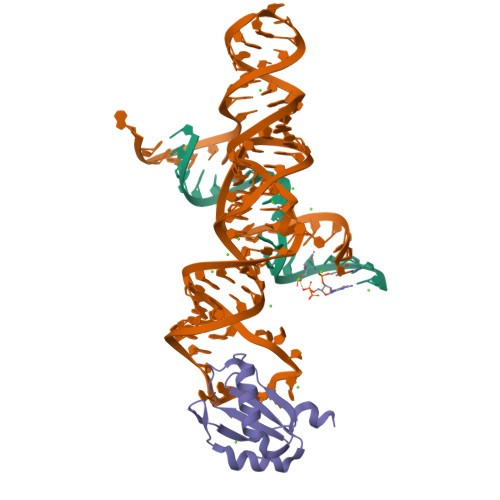
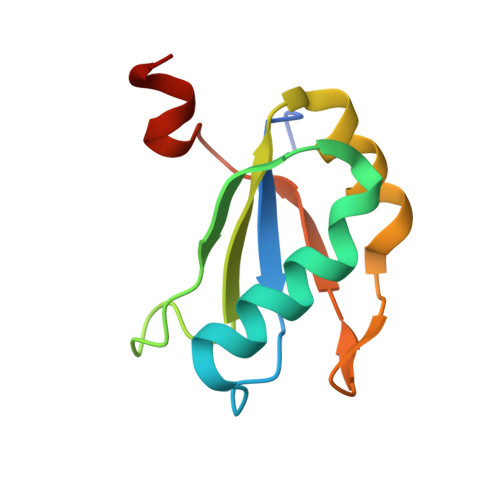
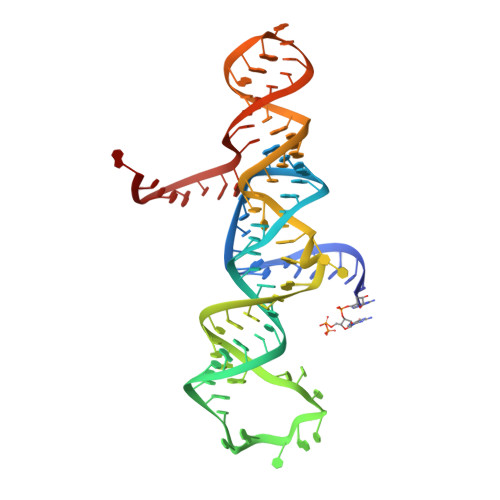
The hairpin ribozyme catalyses sequence-specific cleavage of RNA. The active site of this natural RNA results from the docking of two irregular helices: stems A and B. One strand of stem A harbours the scissile bond. The 2.4 A resolution structure of a hairpin ribozyme-inhibitor complex reveals that the ribozyme aligns the 2'-OH nucleophile and the 5'-oxo leaving group by twisting apart the nucleotides that flank the scissile phosphate. The base of the nucleotide preceding the cleavage site is stacked within stem A; the next nucleotide, a conserved guanine, is extruded from stem A and accommodated by a highly complementary pocket in the minor groove of stem B. Metal ions are absent from the active site. The bases of four conserved purines are positioned potentially to serve as acid-base catalysts. This is the first structure determination of a fully assembled ribozyme active site that catalyses a phosphodiester cleavage without recourse to metal ions.
Organizational Affiliation:
Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington 98109-1024, USA.