Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme.
Wang, L.K., Lima, C.D., Shuman, S.(2002) EMBO J 21: 3873-3880
- PubMed: 12110598
- DOI: https://doi.org/10.1093/emboj/cdf397
- Primary Citation of Related Structures:
1LY1 - PubMed Abstract:
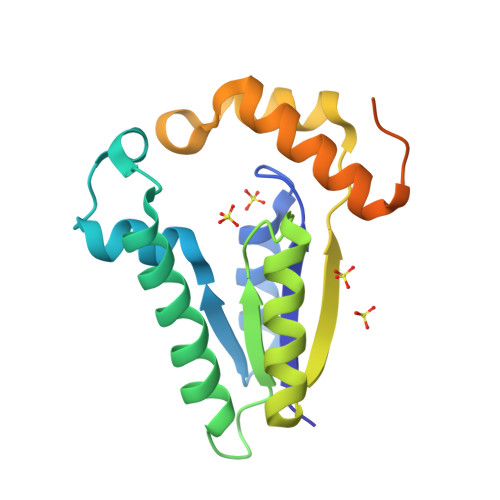
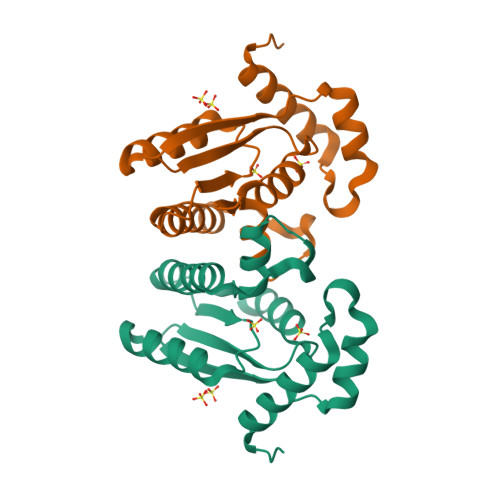
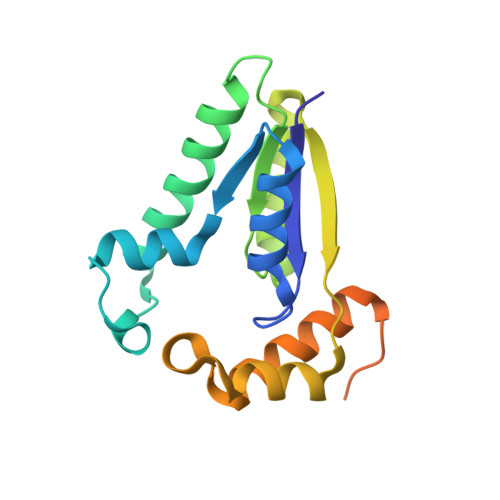
T4 polynucleotide kinase (Pnk), in addition to being an invaluable research tool, exemplifies a family of bifunctional enzymes with 5'-kinase and 3'-phosphatase activities that play key roles in RNA and DNA repair. T4 Pnk is a homotetramer composed of a C-terminal phosphatase domain and an N-terminal kinase domain. The 2.0 A crystal structure of the isolated kinase domain highlights a tunnel-like active site through the heart of the enzyme, with an entrance on the 5' OH acceptor side that can accommodate a single-stranded polynucleotide. The active site is composed of essential side chains that coordinate the beta phosphate of the NTP donor and the 3' phosphate of the 5' OH acceptor, plus a putative general acid that activates the 5' OH. The structure rationalizes the different specificities of T4 and eukaryotic Pnk and suggests a model for the assembly of the tetramer.
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, NY 10021, USA.