Structural Basis for the Molecular Properties of Cytochrome C(6)
Dikiy, A., Carpentier, W., Vandenberghe, I., Borsari, M., Safarov, N., Dikaya, E., Van Beeumen, J., Ciurli, S.(2002) Biochemistry 41: 14689-14699
- PubMed: 12475218
- DOI: https://doi.org/10.1021/bi026473v
- Primary Citation of Related Structures:
1LS9 - PubMed Abstract:
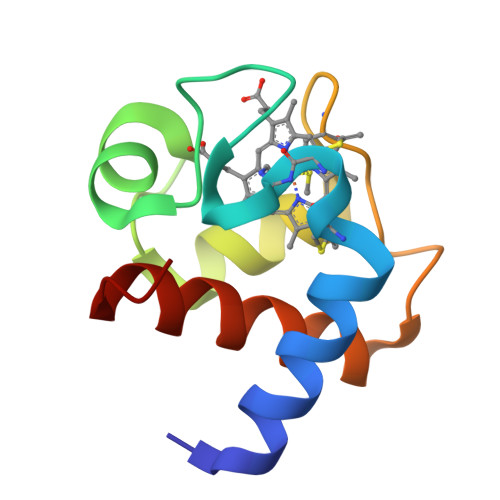
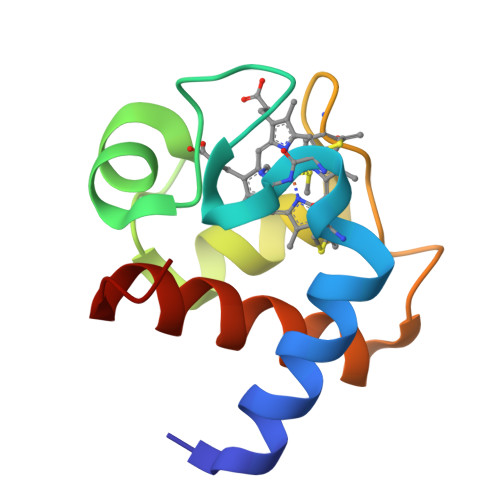
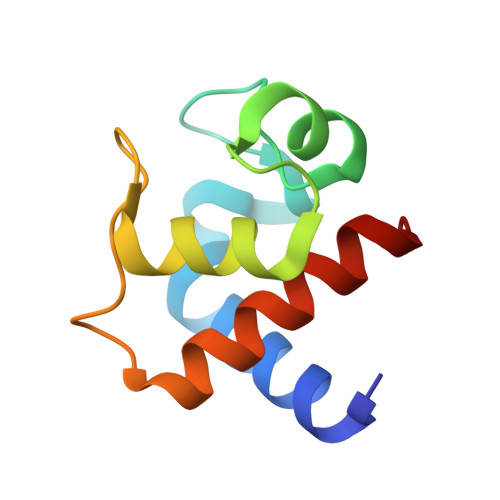
This is a thorough biochemical, spectroscopic, electrochemical, and structural study of a cytochrome c(6) isolated from the filamentous green alga Cladophora glomerata. The protein sequence, elucidated using chemical and mass spectrometric techniques, features 91 amino acids and the characteristic CXXCH heme-binding motif found in c-type cytochromes. The protein is monomeric in both oxidation forms, thereby putting in question a functional role for protein dimerization. Direct electrochemical measurements established, for the first time, the kinetic and thermodynamic data for the redox process in a cytochrome c(6). In particular, the quasi-reversible and diffusion-controlled redox process is accompanied by negative enthalpy and entropy changes, resulting in an E degrees ' value of 0.352 V at 298 K. The pH-dependent properties of the oxidized protein, detected by UV-visible, NMR, and direct cyclic voltammetry, indicate the presence of two acid-base equilibria occurring in the acidic (pK(a) = 4.5) and alkaline regions (pK(a) = 9.0). NMR and electronic spectra allowed the assignment of these equilibria to deprotonation of heme propionate-7 and to replacement of the axial methionine with another ligand, respectively. The 1.3 A resolution X-ray structure of the oxidized protein, revealing a fold typical for class I cytochromes, suggests that the conserved Lys60 replaces the axial methionine at pH >9. The heme solvent accessibility is low, and no water molecules were found in the vicinity of the axial ligands of the heme Fe. A structure-based alignment of cytochromes c(6), and the direct comparison of their structures, indicate a substantial degree of identity between the tertiary structures and suggest patches involved in protein-protein interaction. In particular, the surface electrostatic potential of cytochromes c(6) features a hydrophobic region around the heme cofactor, and a backside surface rich in negative charges.
Organizational Affiliation:
Department of Agro-Environmental Science and Technology, University of Bologna, Via Filippo Re 8, Italy.