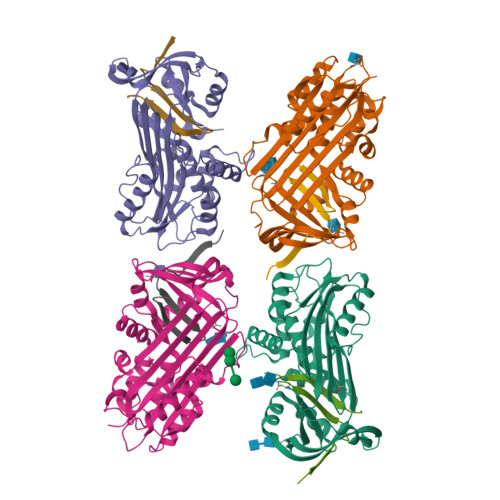
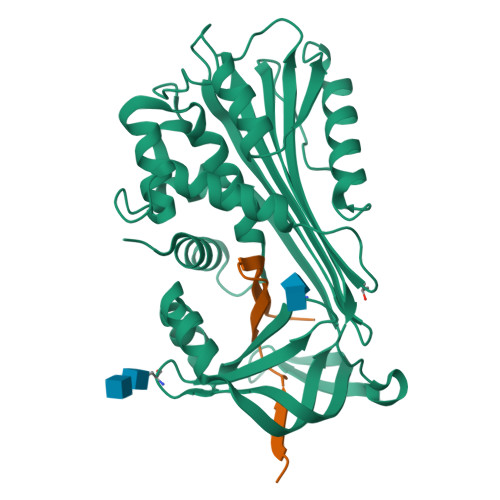
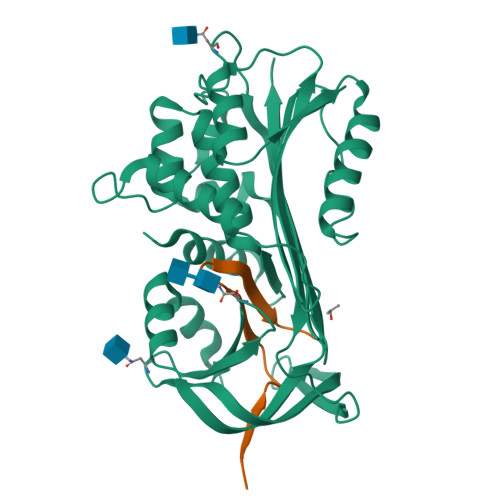
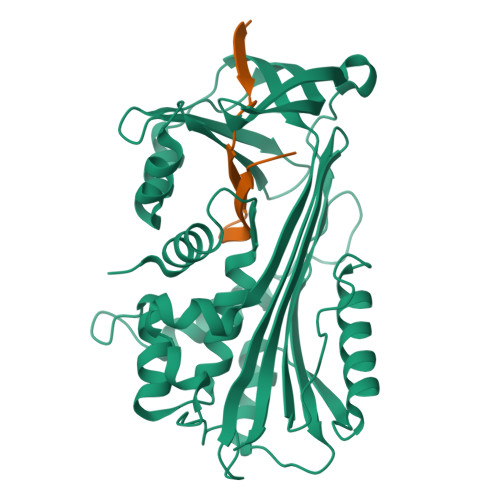
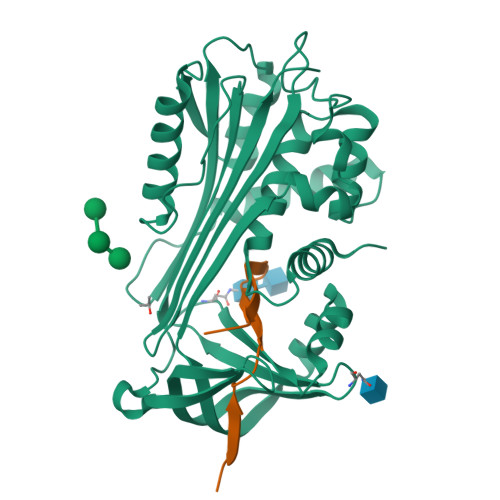
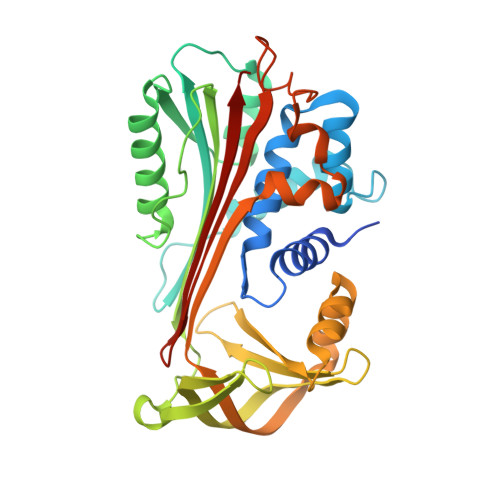
Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Huntington, J.A., Kjellberg, M., Stenflo, J.(2003) Structure 11: 205-211
- PubMed: 12575940
- DOI: https://doi.org/10.1016/s0969-2126(02)00944-9
- Primary Citation of Related Structures:
1LQ8 - PubMed Abstract:
Protein C inhibitor (PCI) is a member of the serpin family that has many biological functions. In blood it acts as a procoagulant, and, in the seminal vesicles, it is required for spermatogenesis. The activity of PCI is affected by heparin binding in a manner unique among the heparin binding serpins, and, in addition, PCI binds hydrophobic hormones with apparent specificity for retinoids. Here we present the 2.4 A crystallographic structure of reactive center loop (RCL) cleaved PCI. A striking feature of the structure is a two-turn N-terminal shortening of helix A, which creates a large hydrophobic pocket that docking studies indicate to be the retinoid binding site. On the basis of surface electrostatic properties, a novel mechanism for heparin activation is proposed.
Organizational Affiliation:
Department of Haematology, Division of Structural Medicine, University of Cambridge, Cambridge Institute for Medical Research, Wellcome Trust/MRC Building, Hills Road, CB2 2XY, Cambridge, United Kingdom. jah52@cam.ac.uk