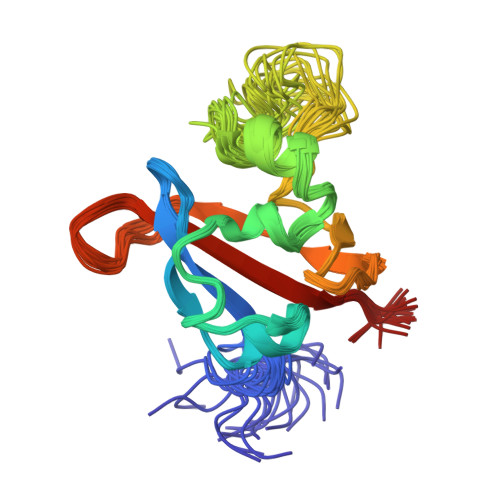
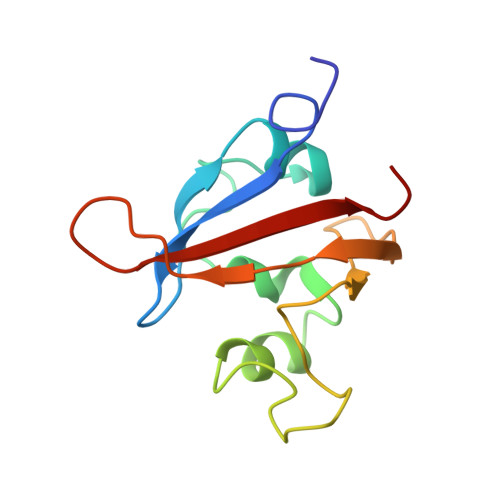
Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation.
Amezcua, C.A., Harper, S.M., Rutter, J., Gardner, K.H.(2002) Structure 10: 1349-1361
- PubMed: 12377121
- DOI: https://doi.org/10.1016/s0969-2126(02)00857-2
- Primary Citation of Related Structures:
1LL8 - PubMed Abstract:
PAS domains are sensory modules in signal-transducing proteins that control responses to various environmental stimuli. To examine how those domains can regulate a eukaryotic kinase, we have studied the structure and binding interactions of the N-terminal PAS domain of human PAS kinase using solution NMR methods. While this domain adopts a characteristic PAS fold, two regions are unusually flexible in solution. One of these serves as a portal that allows small organic compounds to enter into the core of the domain, while the other binds and inhibits the kinase domain within the same protein. Structural and functional analyses of point mutants demonstrate that the compound and ligand binding regions are linked, suggesting that the PAS domain serves as a ligand-regulated switch for this eukaryotic signaling system.
Organizational Affiliation:
Department of Biochemistry, The University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.