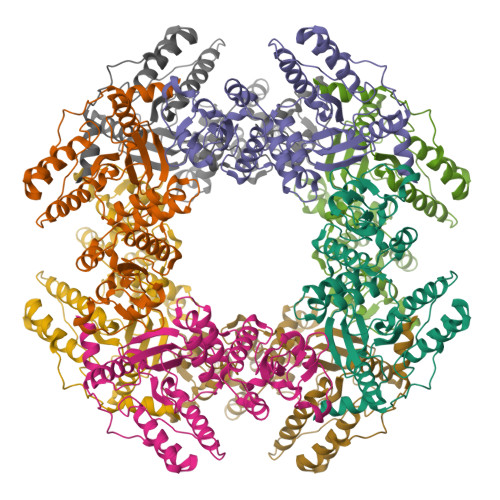
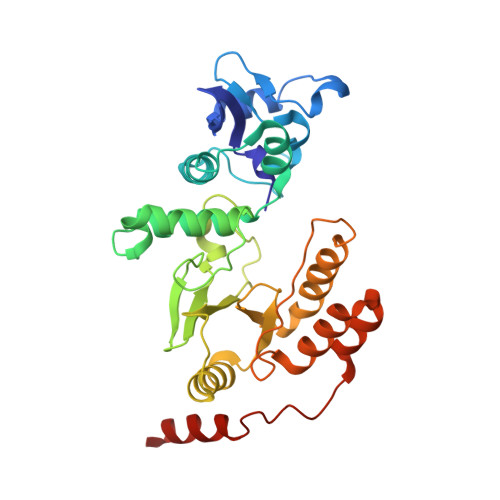
Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold.
Jayaram, H., Taraporewala, Z., Patton, J.T., Prasad, B.V.(2002) Nature 417: 311-315
- PubMed: 12015608
- DOI: https://doi.org/10.1038/417311a
- Primary Citation of Related Structures:
1L9V - PubMed Abstract:
Rotavirus, the major cause of life-threatening infantile gastroenteritis, is a member of the Reoviridae. Although the structures of rotavirus and other members of the Reoviridae have been extensively studied, little is known about the structures of virus-encoded non-structural proteins that are essential for genome replication and packaging. The non-structural protein NSP2 of rotavirus, which exhibits nucleoside triphosphatase, single-stranded RNA binding, and nucleic-acid helix-destabilizing activities, is a major component of viral replicase complexes. We present here the X-ray structure of the functional octamer of NSP2 determined to a resolution of 2.6 A. The NSP2 monomer has two distinct domains. The amino-terminal domain has a new fold. The carboxy-terminal domain resembles the ubiquitous cellular histidine triad (HIT) group of nucleotidyl hydrolases. This structural similarity suggests that the nucleotide-binding site is located inside the cleft between the two domains. Prominent grooves that run diagonally across the doughnut-shaped octamer are probable locations for RNA binding. Several RNA binding sites, resulting from the quaternary organization of NSP2 monomers, may be required for the helix destabilizing activity of NSP2 and its function during genome replication and packaging.
Organizational Affiliation:
Program in Structural and Computational Biology and Molecular Biophysics, Verna and Marrs McLean Department of Biochemistry, Baylor College of Medicine, Houston, Texas 77030, USA.