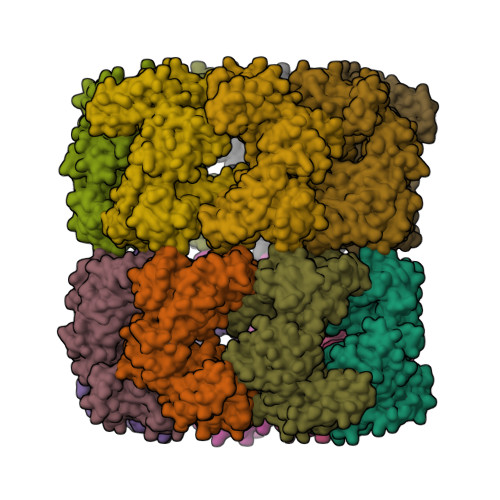
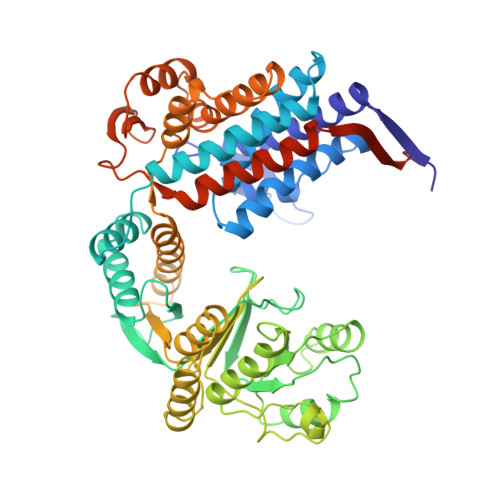
Structural Basis for GroEL-assisted Protein Folding from the Crystal Structure of (GroEL-KMgATP)14 at 2.0 A Resolution
Wang, J., Boisvert, D.C.(2003) J Mol Biol 327: 843-855
- PubMed: 12654267
- DOI: https://doi.org/10.1016/s0022-2836(03)00184-0
- Primary Citation of Related Structures:
1KP8 - PubMed Abstract:
Nucleotide regulates the affinity of the bacterial chaperonin GroEL for protein substrates. GroEL binds protein substrates with high affinity in the absence of ATP and with low affinity in its presence. We report the crystal structure of (GroEL-KMgATP)(14) refined to 2.0 A resolution in which the ATP triphosphate moiety is directly coordinated by both K(+) and Mg(2+). Upon the binding of KMgATP, we observe previously unnoticed domain rotations and a 102 degrees rotation of the apical domain surface helix I. Two major consequences are a large lateral displacement of, and a dramatic reduction of hydrophobicity in, the apical domain surface. These results provide a basis for the nucleotide-dependent regulation of protein substrate binding and suggest a mechanism for GroEL-assisted protein folding by forced unfolding.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry Yale University, 266 Whitney Avenue, Bass Center, Room 418, New Haven, CT 06520-8114, USA. wang@mail.csb.yale.edu