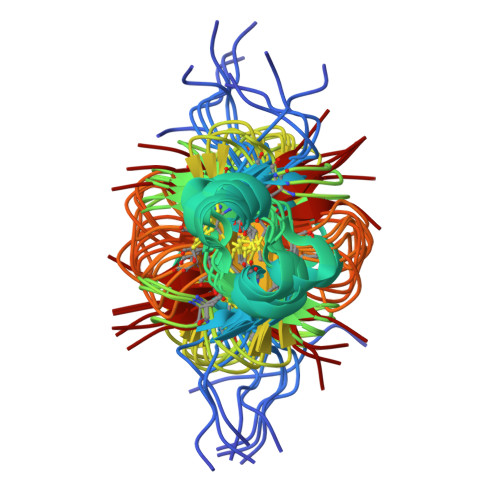
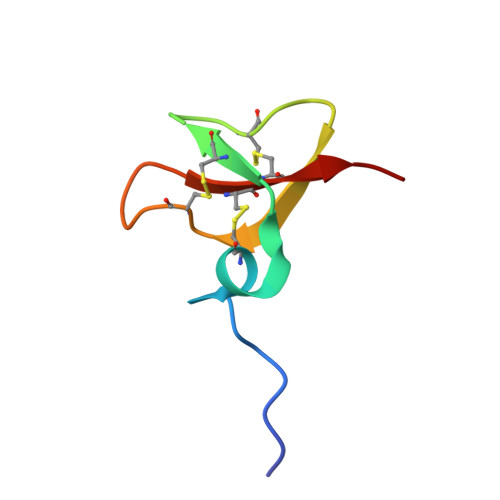
The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
Schibli, D.J., Hunter, H.N., Aseyev, V., Starner, T.D., Wiencek, J.M., McCray Jr., P.B., Tack, B.F., Vogel, H.J.(2002) J Biological Chem 277: 8279-8289
- PubMed: 11741980
- DOI: https://doi.org/10.1074/jbc.M108830200
- Primary Citation of Related Structures:
1KJ5, 1KJ6 - PubMed Abstract:
The three human beta-defensins, HBD1--3, are 33--47-residue, cationic antimicrobial proteins expressed by epithelial cells. All three proteins have broad spectrum antimicrobial activity, with HBD3 consistently being the most potent. Additionally, HBD3 has significant bactericidal activity against Gram-positive Staphylococcus aureus at physiological salt concentrations. We have compared the multimeric state of the three beta-defensins using NMR diffusion spectroscopy, dynamic and static light scattering, and analysis of the migration of the three beta-defensins on a native gel. All three techniques are in agreement, suggesting that HBD-3 is a dimer, while HBD-1 and HBD-2 are monomeric. Subsequently, the NMR solution structures of HBD1 and HBD3 were determined using standard homonuclear techniques and compared with the previously determined solution structure of HBD2. Both HBD1 and HBD3 form well defined structures with backbone root mean square deviations of 0.451 and 0.616 A, respectively. The tertiary structures of all three beta-defensins are similar, with a short helical segment preceding a three-stranded antiparallel beta-sheet. The surface charge density of each of the defensins is markedly different, with the surface of HBD3 significantly more basic. Analysis of the NMR data and structures led us to suggest that HBD3 forms a symmetrical dimer through strand beta2 of the beta-sheet. The increased anti-Staphylococcal activity of HBD3 may be explained by the capacity of the protein to form dimers in solution at low concentrations, an amphipathic dimer structure, and the increased positive surface charge compared with HBD1 and HBD2.
Organizational Affiliation:
Structural Biology Research Group, Department of Biological Sciences, University of Calgary, Calgary, Alberta T2N 1N4, Canada.