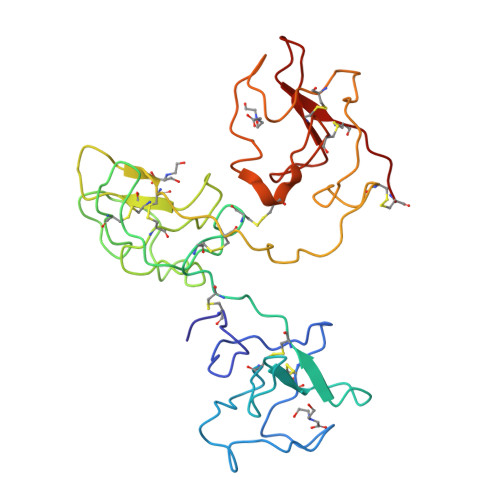
The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin.
Abad, M.C., Arni, R.K., Grella, D.K., Castellino, F.J., Tulinsky, A., Geiger, J.H.(2002) J Mol Biol 318: 1009-1017
- PubMed: 12054798
- DOI: https://doi.org/10.1016/S0022-2836(02)00211-5
- Primary Citation of Related Structures:
1KI0 - PubMed Abstract:
Angiogenesis inhibitors have gained much public attention recently as anti-cancer agents and several are currently in clinical trials, including angiostatin (Phase I, Thomas Jefferson University Hospital, Philadelphia, PA). We report here the bowl-shaped structure of angiostatin kringles 1-3, the first multi-kringle structure to be determined. All three kringle lysine-binding sites contain a bound bicine molecule of crystallization while the former of kringle 2 and kringle 3 are cofacial. Moreover, the separation of the kringle 2 and kringle 3 lysiner binding sites is sufficient to accommodate the alpha-helix of the 30 residue peptide VEK-30 found in the kringle 2/VEK-30 complex. Together the three kringles produce a central cavity suggestive of a unique domain where they may function in concert.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing 48824, USA.