Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains.
Misra, S., Puertollano, R., Kato, Y., Bonifacino, J.S., Hurley, J.H.(2002) Nature 415: 933-937
- PubMed: 11859375
- DOI: https://doi.org/10.1038/415933a
- Primary Citation of Related Structures:
1JPL, 1JUQ - PubMed Abstract:
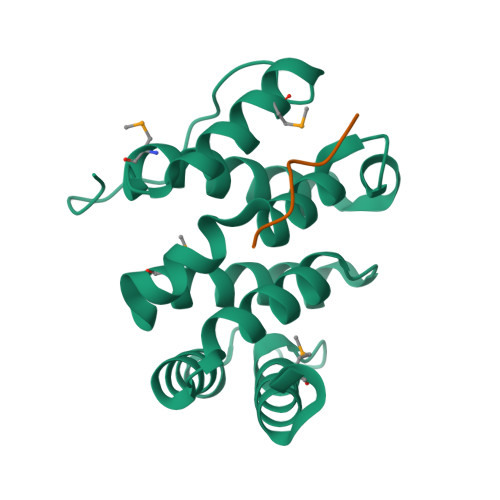
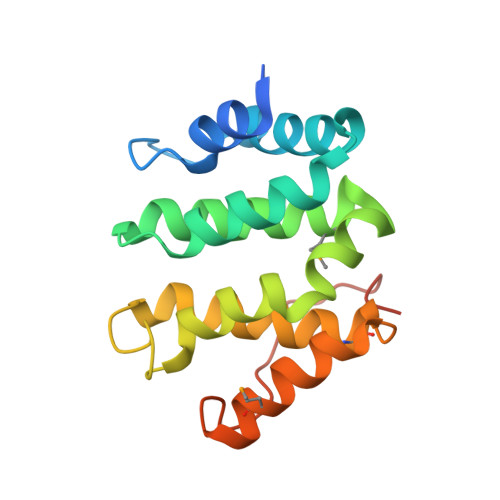
Specific sorting signals direct transmembrane proteins to the compartments of the endosomal-lysosomal system. Acidic-cluster-dileucine signals present within the cytoplasmic tails of sorting receptors, such as the cation-independent and cation-dependent mannose-6-phosphate receptors, are recognized by the GGA (Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding) proteins. The VHS (Vps27p, Hrs and STAM) domains of the GGA proteins are responsible for the highly specific recognition of these acidic-cluster-dileucine signals. Here we report the structures of the VHS domain of human GGA3 complexed with signals from both mannose-6-phosphate receptors. The signals bind in an extended conformation to helices 6 and 8 of the VHS domain. The structures highlight an Asp residue separated by two residues from a dileucine sequence as critical recognition elements. The side chains of the Asp-X-X-Leu-Leu sequence interact with subsites consisting of one electropositive and two shallow hydrophobic pockets, respectively. The rigid spatial alignment of the three binding subsites leads to high specificity.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.