Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges.
Satoh, A., Kim, J.K., Miyahara, I., Devreese, B., Vandenberghe, I., Hacisalihoglu, A., Okajima, T., Kuroda, S., Adachi, O., Duine, J.A., Van Beeumen, J., Tanizawa, K., Hirotsu, K.(2002) J Biol Chem 277: 2830-2834
- PubMed: 11704672
- DOI: https://doi.org/10.1074/jbc.M109090200
- Primary Citation of Related Structures:
1JMX, 1JMZ - PubMed Abstract:
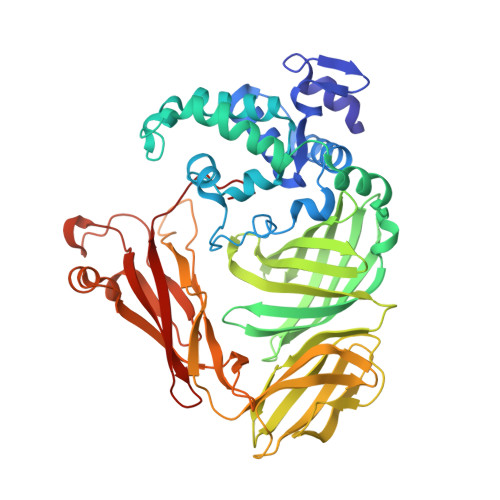
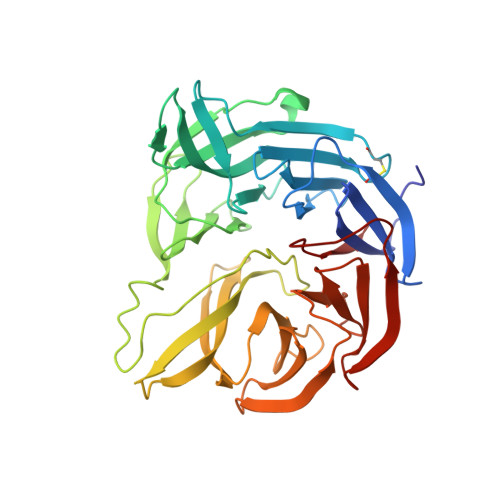
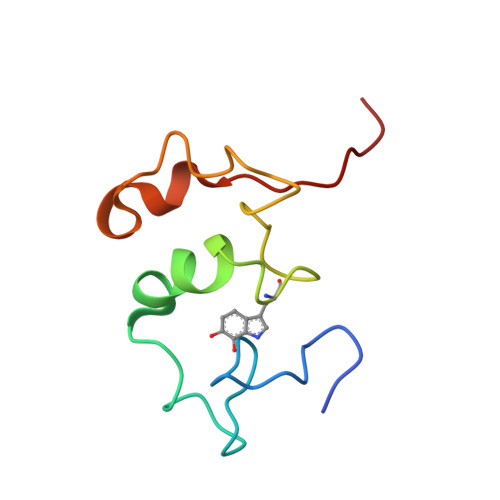
The crystal structure of a quinohemoprotein amine dehydrogenase from Pseudomonas putida has been determined at 1.9-A resolution. The enzyme comprises three non-identical subunits: a four-domain alpha-subunit that harbors a di-heme cytochrome c, a seven-bladed beta-propeller beta-subunit that provides part of the active site, and a small gamma-subunit that contains a novel cross-linked, proteinous quinone cofactor, cysteine tryptophylquinone. More surprisingly, the catalytic gamma-subunit contains three additional chemical cross-links that encage the cysteine tryptophylquinone cofactor, involving a cysteine side chain bridged to either an Asp or Glu residue all in a hitherto unknown thioether bonding with a methylene carbon atom of acidic amino acid side chains. Thus, the structure of the 79-residue gamma-subunit is quite unusual, containing four internal cross-links in such a short polypeptide chain that would otherwise be difficult to fold into a globular structure.
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Osaka City University, Osaka 558-8585, Japan.