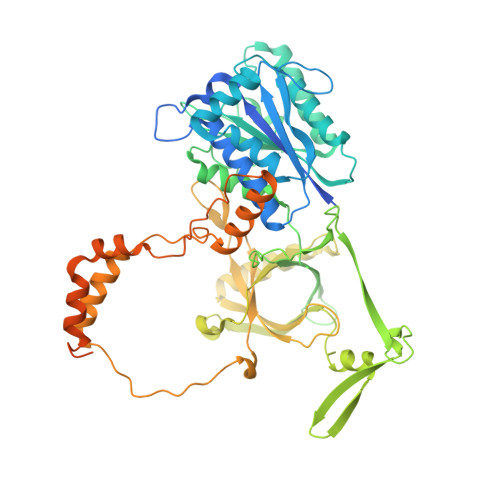
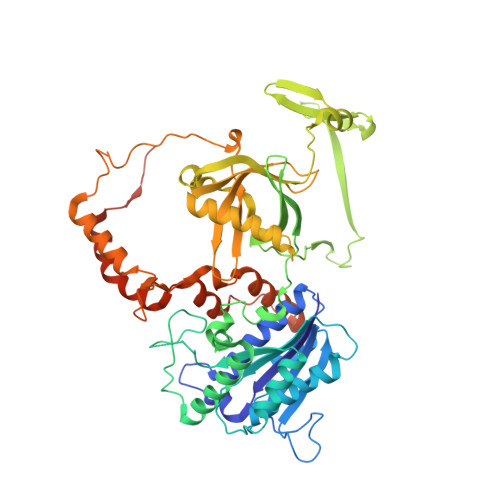
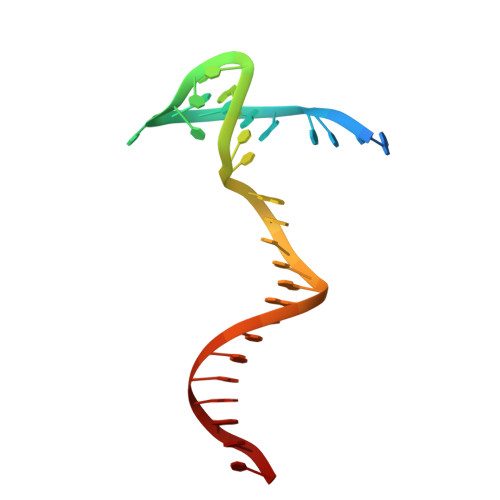
Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair.
Walker, J.R., Corpina, R.A., Goldberg, J.(2001) Nature 412: 607-614
- PubMed: 11493912
- DOI: https://doi.org/10.1038/35088000
- Primary Citation of Related Structures:
1JEQ, 1JEY - PubMed Abstract:
The Ku heterodimer (Ku70 and Ku80 subunits) contributes to genomic integrity through its ability to bind DNA double-strand breaks and facilitate repair by the non-homologous end-joining pathway. The crystal structure of the human Ku heterodimer was determined both alone and bound to a 55-nucleotide DNA element at 2.7 and 2.5 A resolution, respectively. Ku70 and Ku80 share a common topology and form a dyad-symmetrical molecule with a preformed ring that encircles duplex DNA. The binding site can cradle two full turns of DNA while encircling only the central 3-4 base pairs (bp). Ku makes no contacts with DNA bases and few with the sugar-phosphate backbone, but it fits sterically to major and minor groove contours so as to position the DNA helix in a defined path through the protein ring. These features seem well designed to structurally support broken DNA ends and to bring the DNA helix into phase across the junction during end processing and ligation.
Organizational Affiliation:
Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10021, USA.