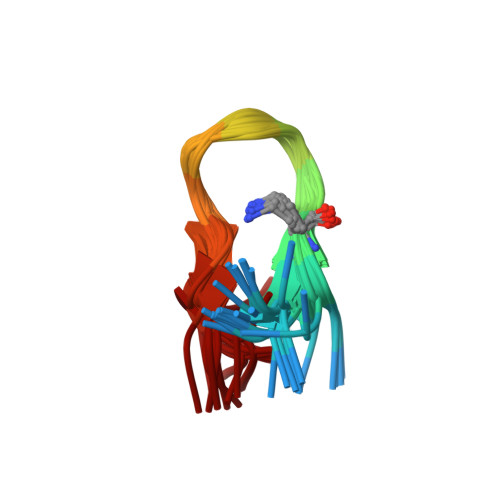
Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
Du, A.P., Limal, D., Semetey, V., Dali, H., Jolivet, M., Desgranges, C., Cung, M.T., Briand, J.P., Petit, M.C., Muller, S.(2002) J Mol Biology 323: 503-521
- PubMed: 12381305
- DOI: https://doi.org/10.1016/s0022-2836(02)00701-5
- Primary Citation of Related Structures:
1IM7, 1J8N, 1J8Z, 1J9V, 1JAA, 1JAR, 1JC8, 1JCP, 1JD8, 1JDK - PubMed Abstract:
The conformational and immunological properties of different analogues corresponding to the 600-612 disulfide loop of the human immunodeficiency virus (HIV) gp41 glycoprotein envelope were studied. Fourteen analogues were designed and synthesised; namely, a series of seven analogues in which the disulfide bond was replaced by a lactam bridge and a series of seven analogues in which one residue of each analogue at a time, was replaced by its corresponding homologised alpha-amino acid (beta(3)-amino acid). In the case of the lactam analogues, the influence of the two possible CO-NH and NH-CO orientations of the lactam bridge as well as the size of the lactam ring was explored. The analogues were tested in ELISA with monoclonal antibodies raised against the 600-612 cyclic parent peptide as well as with sera from HIV-1 infected patients. A structural analysis of the parent and analogue peptides was carried out in dimethyl sulfoxide (DMSO-d(6)) using two-dimensional NMR techniques and molecular dynamics simulations. Comparison of the own conformation of the cyclic analogues with their either strong or weak reactivity with the antibodies reveals structural features that may be correlated with the antibody reactivity. Thus, a close structural similarity, particularly a characteristic orientation of the side-chains of residues Lys606, Leu607 and Ile608 in the loop, was found in certain beta(3)-analogues that were better recognised than the parent peptide by anti-peptide mouse monoclonal antibodies and patients' antibodies.
Organizational Affiliation:
Laboratoire de Chimie Physique Macromoléculaire, Unité Mixte de Recherches 7568 CNRS-INPL, 54000, Nancy, France.