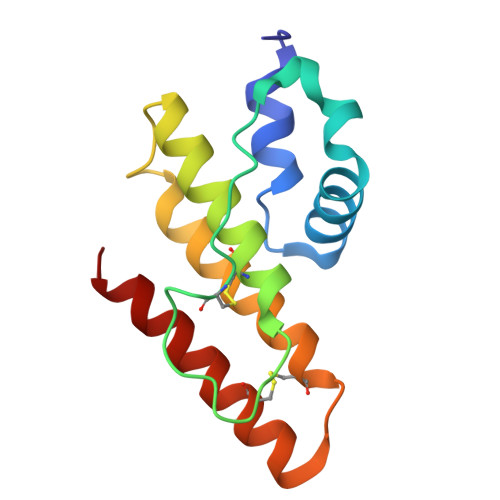
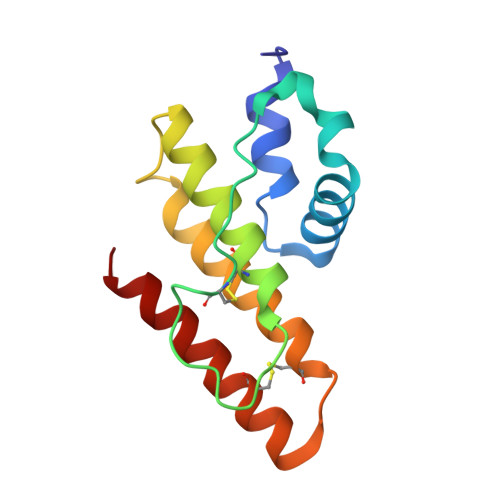
A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
Sugiyama, M., Ohtani, K., Izuhara, M., Koike, T., Suzuki, K., Imamura, S., Misaki, H.(2002) J Biological Chem 277: 20051-20058
- PubMed: 11897786
- DOI: https://doi.org/10.1074/jbc.M200264200
- Primary Citation of Related Structures:
1IT4, 1IT5 - PubMed Abstract:
Until now, phospholipase A(2) (PLA(2); EC 3.1.14) has been found only from eukaryotic sources. In the present study, we found a secreted PLA(2), which is produced by a soil bacterium, Streptomyces violaceoruber A-2688, demonstrating that the enzyme is the first phospholipase A(2) identified in prokaryote. After characterization of the novel PLA(2), a gene encoding the enzyme was cloned, sequenced, and overexpressed using a Streptomyces host-vector system. The amino acid sequence showed that the prokaryotic PLA(2) has only four cysteines and less homology to the eukaryotic ones, which have 12-16 cysteines. The solution structures of the prokaryotic PLA(2), bound and unbound with calcium(II) ion, were determined by using the NMR technique and structure calculation. The overall structure of the S. violaceoruber PLA(2), which is composed of only five alpha-helices, is completely different from those of eukaryotic PLA(2)s, which consist of beta-sheets and alpha-helices. The structure of the calcium-binding domain is obviously distinct from that without the ion; the ligands for the calcium(II) ion are the two carboxylates of Asp(43) (monodentate) and Asp(65) (bidentate), the carbonyl oxygen of Leu(44), and three water molecules. A calcium-binding experiment showed that the calcium dissociation constant ( approximately 5 mm) for the prokaryotic PLA(2) is much larger than those of eukaryotic ones.
Organizational Affiliation:
Institute of Pharmaceutical Sciences, Faculty of Medicine, Hiroshima University, Kasumi 1-2-3, Minami-ku, Hiroshima 734-8551, Japan.