Structure of aspergillopepsin I from Aspergillus phoenicis: variations of the S1'-S2 subsite in aspartic proteinases.
Cho, S.W., Kim, N., Choi, M.U., Shin, W.(2001) Acta Crystallogr D Biol Crystallogr 57: 948-956
- PubMed: 11418762
- DOI: https://doi.org/10.1107/s0907444901005972
- Primary Citation of Related Structures:
1IBQ - PubMed Abstract:
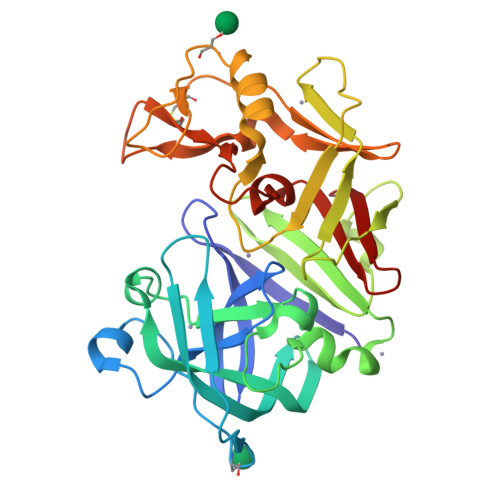
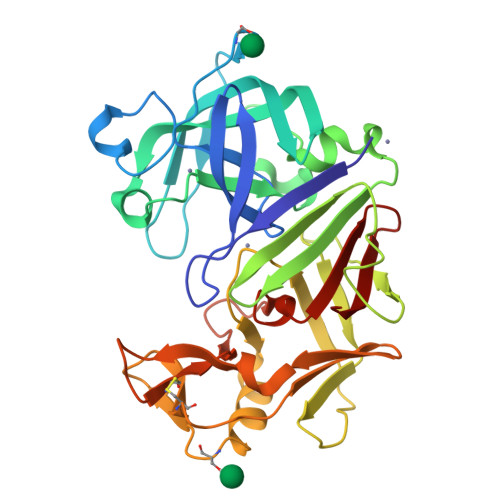
The crystal structure of aspergillopepsin I (AP) from Aspergillus phoenicis has been determined at 2.18 A resolution and refined to R and R(free) factors of 21.5 and 26.0%, respectively. AP has the typical two beta-barrel domain structure of aspartic proteinases. The structures of the two independent molecules are partly different, exemplifying the flexible nature of the aspartic proteinase structure. Notably, the 'flap' in one molecule is closer, with a largest separation of 4.0 A, to the active site than in the other molecule. AP is most structurally homologous to penicillopepsin (PP) and then to endothiapepsin (EP), which share sequence identities of 68 and 56%, respectively. However, AP is similar to EP but differs from PP in the combined S1'-S2 subsite that is delineated by a flexible psi-loop in the C-terminal domain. The S1' and S2 subsites are well defined and small in AP, while there is no definite border between S1' and S2 and the open space for the S2 subsite is larger in PP. Comparison of the structures indicates that the two amino-acid residues equivalent to Leu295 and Leu297 of AP are the major determining factors in shaping the S1'-S2 subsite in the fungal aspartic proteinases.
Organizational Affiliation:
School of Chemistry and Molecular Engineering, Seoul National University, South Korea.