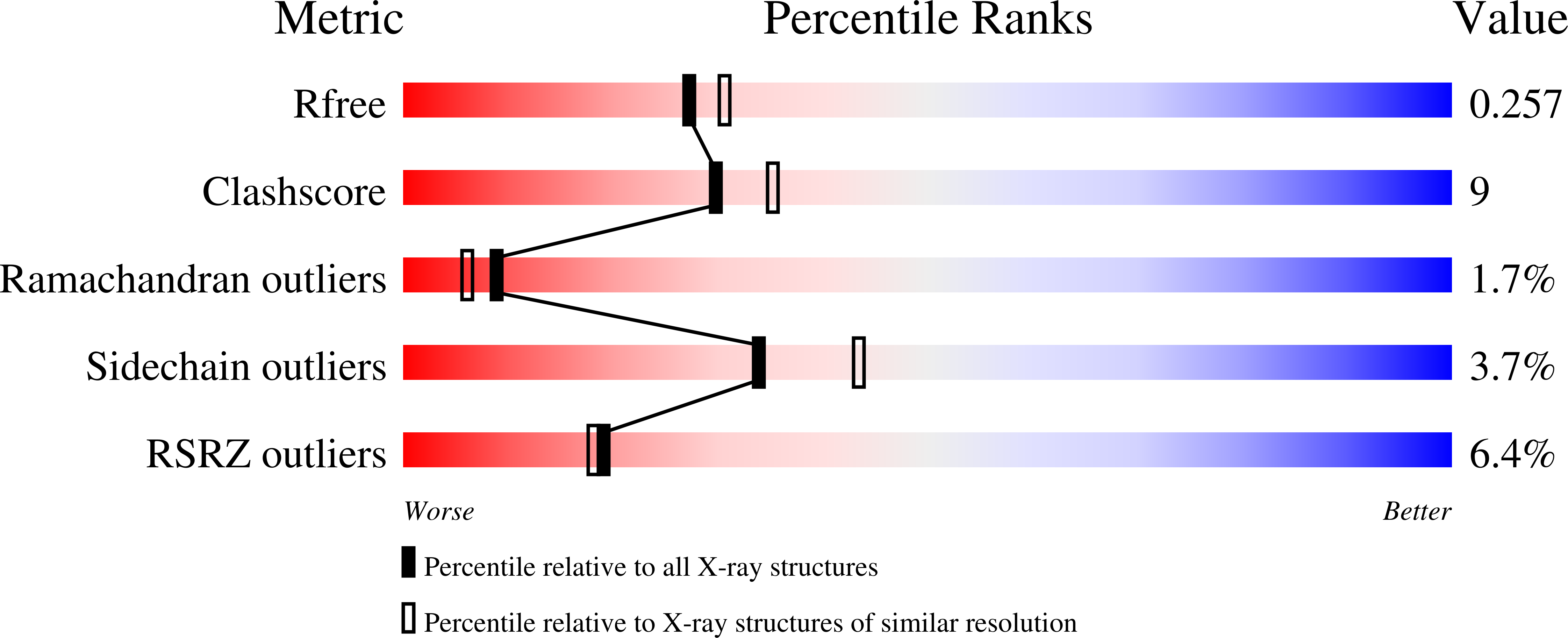
Structure of staphylococcal enterotoxin C2 at various pH levels.
Kumaran, D., Eswaramoorthy, S., Furey, W., Sax, M., Swaminathan, S.(2001) Acta Crystallogr D Biol Crystallogr 57: 1270-1275
- PubMed: 11526318
- DOI: https://doi.org/10.1107/s0907444901011118
- Primary Citation of Related Structures:
1CQV, 1I4P, 1I4Q, 1I4R, 1I4X - PubMed Abstract:
The three-dimensional structure of staphylococcal enterotoxin C2 (SEC2), a toxin as well as a superantigen, has been determined at various pH levels from two different crystal forms, tetragonal (pH 5.0, 5.5, 6.0 and 6.5) and monoclinic (pH 8.0) at 100 and 293 K, respectively, by the molecular-replacement method. Tetragonal crystals belong to space group P4(3)2(1)2, with unit-cell parameters a = b = 42.68, c = 289.15 A (at pH 5.0), and monoclinic crystals to space group P2(1), with unit-cell parameters a = 43.3, b = 70.6, c = 42.2 A, beta = 90.3 degrees. SEC2 contains a zinc-binding motif, D+HExxH, and accordingly a Zn atom has been identified. The coordination of the zinc ion suggests that it may be catalytic zinc rather than structural, but there is so far no biological evidence that it possesses catalytic activity. However, superantigen staphylococcal exfoliative toxins A and B have been shown to have enzymatic activity after their fold was identified to be similar to that of serine protease. The structure and its conformation are similar to the previously reported structures of SEC2. Though it was expected that the zinc ion may be leached out, as the histidines coordinating the zinc ion are expected to be protonated below pH 6.0, zinc is present at all pH values. The coordination distances to zinc increase with decreasing pH, with the distances being the least at pH 8.0. The results of automated model building using the ARP/wARP program for different data sets collected at various pH values are discussed.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA.