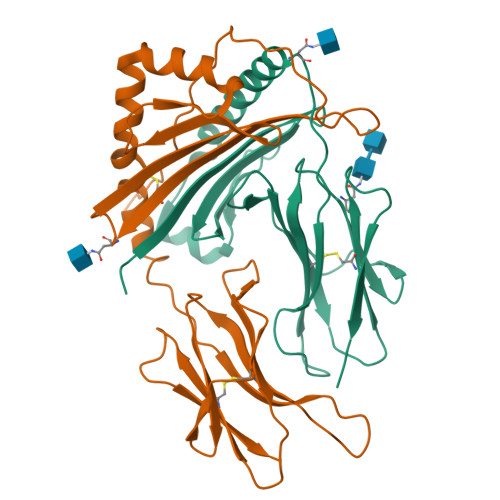
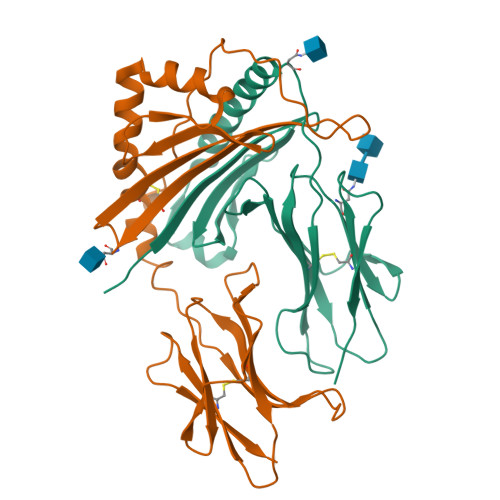
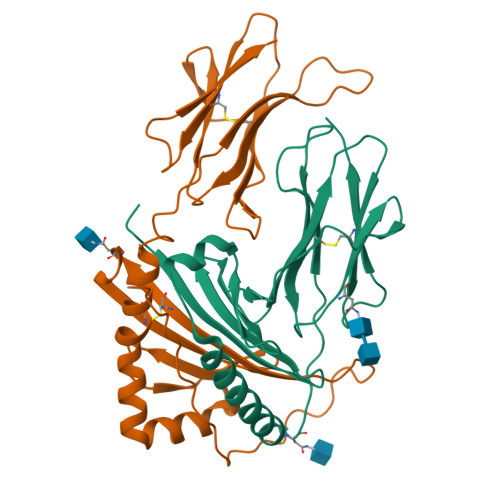
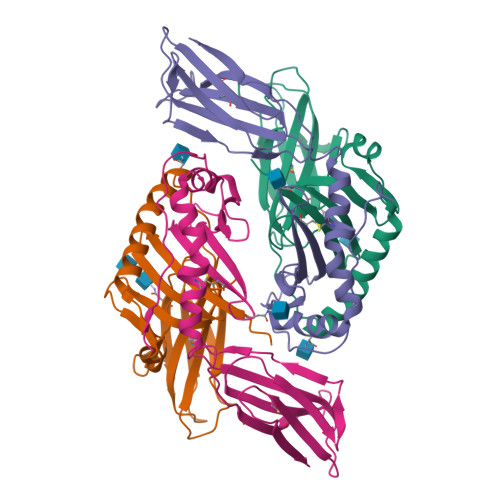
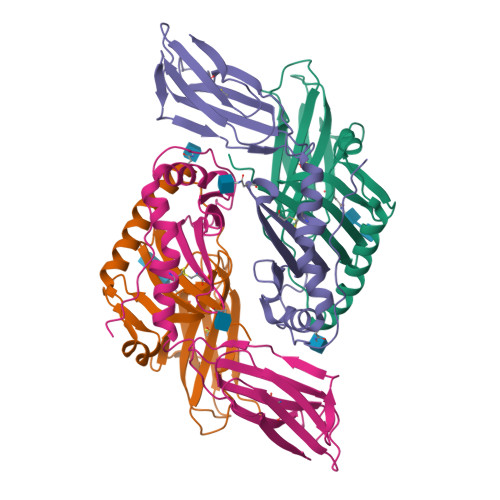
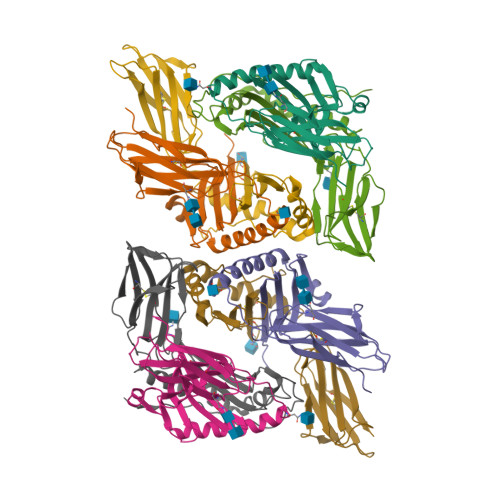Mutations changing the kinetics of class II MHC peptide exchange.
Wilson, N., Fremont, D., Marrack, P., Kappler, J.(2001) Immunity 14: 513-522
- PubMed: 11371354
- DOI: https://doi.org/10.1016/s1074-7613(01)00140-6
- Primary Citation of Related Structures:
1I3R - PubMed Abstract:
IE/DR MHC class II molecules have an extensive H-bonding network under the bound peptide. In IE(k), two alpha chain acidic amino acids in the core of this network were mutated to amides. At low pH, the mutant molecule exchanged peptide much more rapidly than the wild-type. The crystal structure of the mutant IE(k) revealed the loss of a single buried water molecule and a reorganization of the predicted H-bonding network. We suggest that these mutations enhance the transition of MHC class II to an open conformation at low pH allowing the bound peptide to escape. In wild-type IE(k), the need to protonate these amino acids also may be a bottleneck in the return to a closed conformation after peptide binding.
Organizational Affiliation:
Howard Hughes Medical Institute, Integrated Department of Immunology, Zuckerman Family/Canyon, Ranch Crystallography Laboratory, National Jewish Medical and Research Center, Denver, CO 80206, USA.