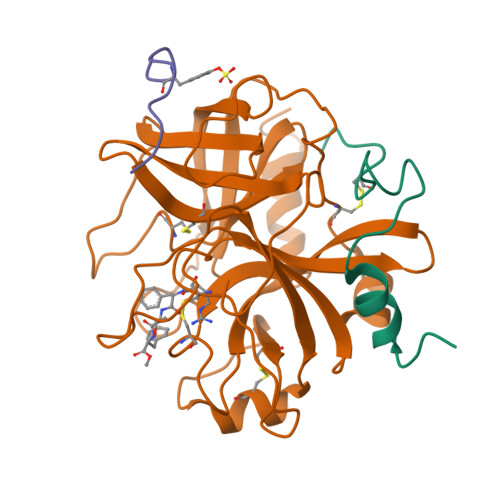
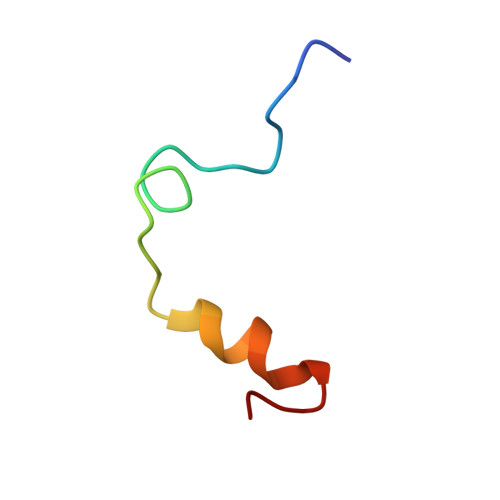
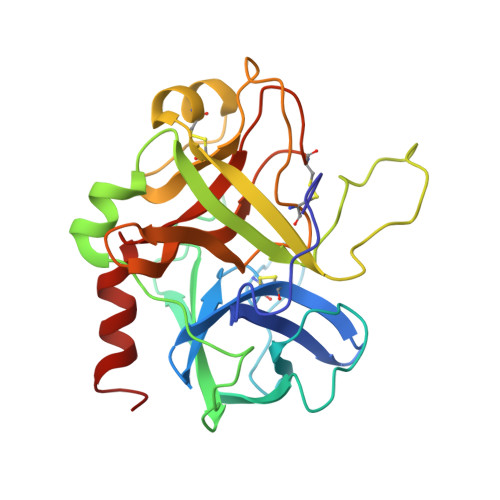
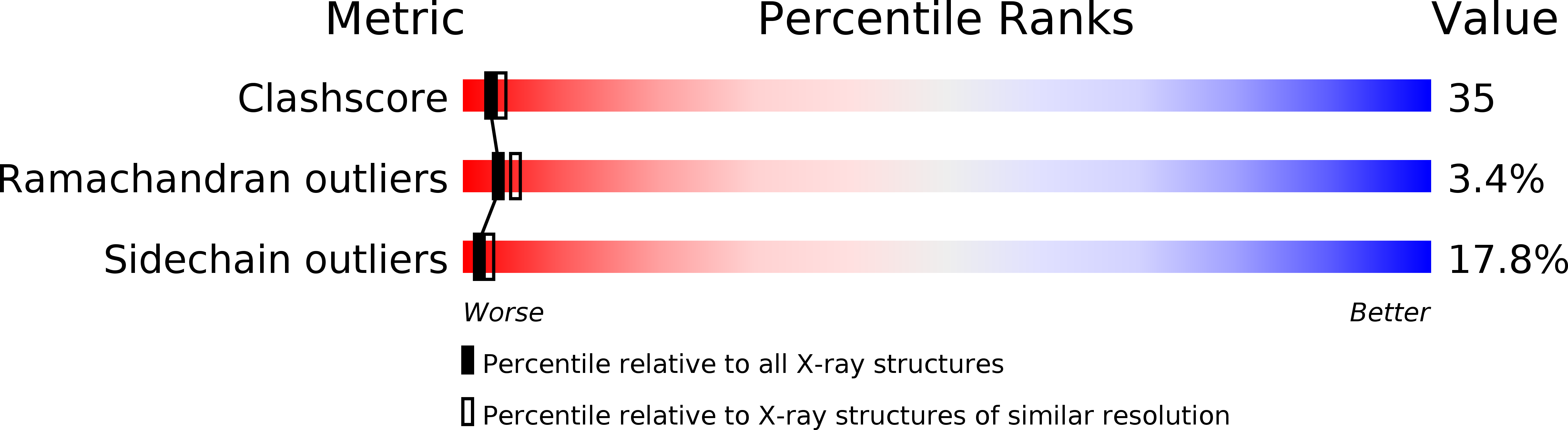Structure of a retro-binding peptide inhibitor complexed with human alpha-thrombin.
Tabernero, L., Chang, C.Y., Ohringer, S.L., Lau, W.F., Iwanowicz, E.J., Han, W.C., Wang, T.C., Seiler, S.M., Roberts, D.G., Sack, J.S.(1995) J Mol Biology 246: 14-20
- PubMed: 7853394
- DOI: https://doi.org/10.1006/jmbi.1994.0060
- Primary Citation of Related Structures:
1HDT - PubMed Abstract:
The crystallographic structure of the ternary complex between human alpha-thrombin, hirugen and the peptidyl inhibitor Phe-alloThr-Phe-O-CH3, which is acylated at its N terminus with 4-guanidino butanoic acid (BMS-183507), has been determined at 2.6 A resolution. The structure reveals a unique "retro-binding" mode for this tripeptide active site inhibitor. The inhibitor binds with its alkyl-guanidine moiety in the primary specificity pocket and its two phenyl rings occupying the hydrophobic proximal and distal pockets of the thrombin active site. In this arrangement the backbone of the tripeptide forms a parallel beta-strand to the thrombin main-chain at the binding site. This is opposite to the orientation of the natural substrate, fibrinogen, and all the small active site-directed thrombin inhibitors whose bound structures have been previously reported. BMS-183507 is the first synthetic inhibitor proved to bind in a retro-binding fashion to thrombin, in a fashion similar to that of the N-terminal residues of the natural inhibitor hirudin. Furthermore, this new potent thrombin inhibitor (Ki = 17.2 nM) is selective for thrombin over other serine proteases tested and may be a template to be considered in designing hirudin-based thrombin inhibitors with interactions at the specificity pocket.
Organizational Affiliation:
Department of Macromolecular Crystallography, Bristol-Myers Squibb Pharmaceutical Research Institute, Princeton, NJ 08543-4000.