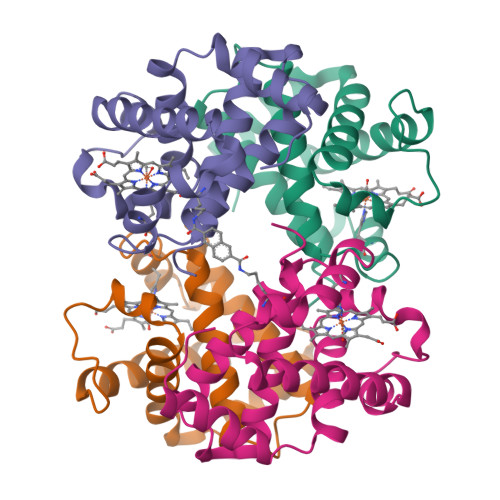
Allosteric intermediates indicate R2 is the liganded hemoglobin end state.
Schumacher, M.A., Zheleznova, E.E., Poundstone, K.S., Kluger, R., Jones, R.T., Brennan, R.G.(1997) Proc Natl Acad Sci U S A 94: 7841-7844
- PubMed: 9223274
- DOI: https://doi.org/10.1073/pnas.94.15.7841
- Primary Citation of Related Structures:
1HAB, 1HAC - PubMed Abstract:
Hemoglobin has been a long-standing paradigm for understanding protein allostery. Here, the x-ray structures of two chemically crosslinked, fully liganded hemoglobins, alpha2beta82CA82beta and alpha2beta82ND82beta, are described at 2.3 A and 2.6 A resolution, respectively. Strikingly, these crosslinked hemoglobins assume intermediate conformations that lie between those of R and the controversial liganded hemoglobin state R2 rather than between R and T. Thus, these structures support only a T left and right arrow R left and right arrow R2 allosteric pathway and underscore the physiological importance of the R2 conformation.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Oregon Health Sciences University, 3181 SW Sam Jackson Park Road, Portland, OR 97201-3098, USA.