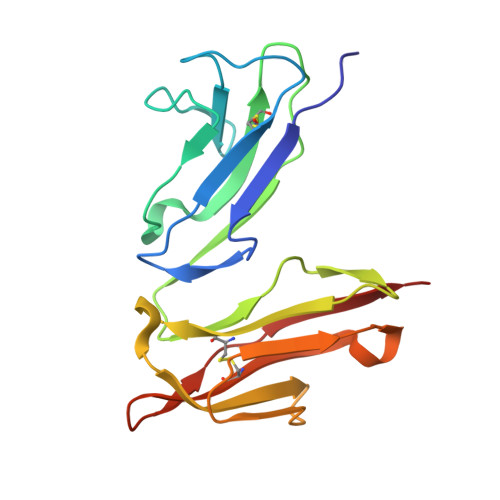
Molecular Basis for Immune Complex Recognition: A Comparison of Fc-Receptor Structures
Sondermann, P., Kaiser, J., Jacob, U.(2001) J Mol Biology 309: 737
- PubMed: 11397093
- DOI: https://doi.org/10.1006/jmbi.2001.4670
- Primary Citation of Related Structures:
1H9V - PubMed Abstract:
Once antigen is opsonised by IgG it is removed from the circulation by Fcgamma-receptor expressing cells. Fcgamma-receptors are type I transmembrane molecules that carry extracellular parts consisting of two or three immunoglobulin domains. Previously solved structures of Fc-receptors reveal that the N-terminal two Ig-like domains are arranged in a steep angle forming a heart-shaped structure. The crystal structure of the FcgammaRIII/hIgG1-Fc-fragment demonstrated that the Fc-fragment is recognised through loops of the C-terminal receptor domain of the FcgammaRIII. As the overall structure of the FcRs and their Ig ligands are very similar we modelled the Ig complexes with FcgammaRI, FcgammaRII and FcepsilonRIalpha based on the FcgammaRIII/hIgG1-Fc-fragment structure. The obtained models are consistent with the observed biochemical data and may explain the observed specificity and affinities.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18a, Martinsried, D-82152, Germany. sonderma@biochem.mpg.de